Workflow Type: KNIME
Open
Stable
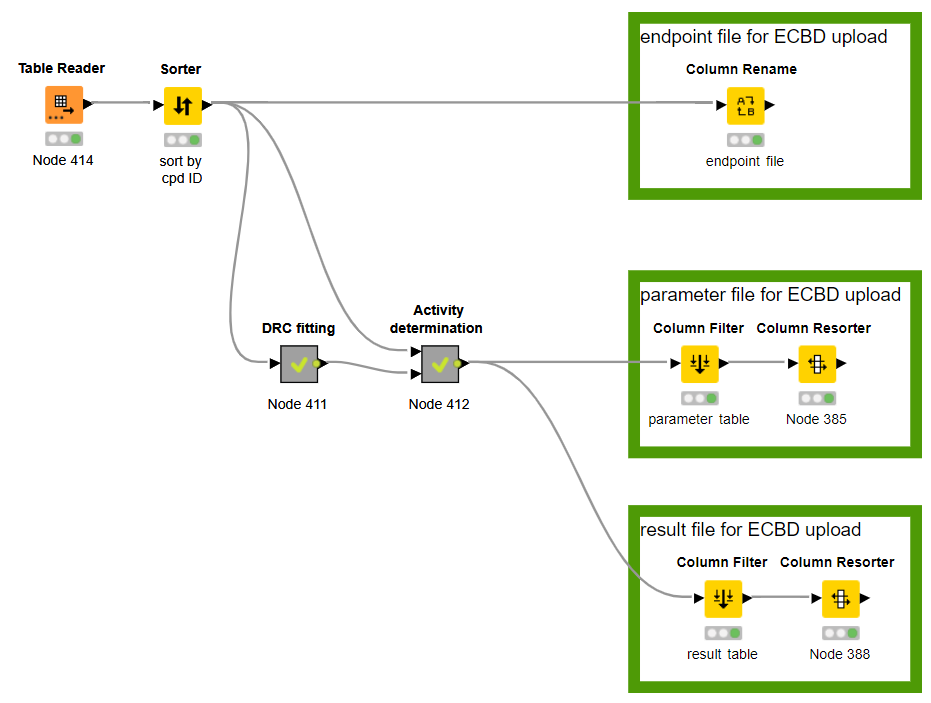
This workflow can be used to fit dose-response curves from normalised biochemical assay data (%Inhibition) using the HCS extension. This workflow needs R-Server to run in the back-end. Start R and run the following command: library(Rserve); Rserve(args = "--vanilla") IC50 values will not be extrapolated outside the tested concentration range For activity classification the following criteria are applied:
- maximum (average % inhibion) >25 % and slope is >0 and IC50 > 5 µM or
- minimum (average % inhibion) >75 % Results are formatted for upload to the European Chemical Biology Database (ECBD)
Version History
Version 1 (earliest) Created 26th Sep 2022 at 11:15 by Jeanette Reinshagen
Initial commit
Open
 master
master9abaa54
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Activity
Views: 2197 Downloads: 359
Created: 26th Sep 2022 at 11:15
Annotated Properties
Topic annotations
 Tags
TagsThis item has not yet been tagged.
 Attributions
AttributionsNone
 Visit source
Visit source https://orcid.org/0000-0002-8080-9170
https://orcid.org/0000-0002-8080-9170