Workflow Type: Snakemake
Frozen
Frozen
Stable
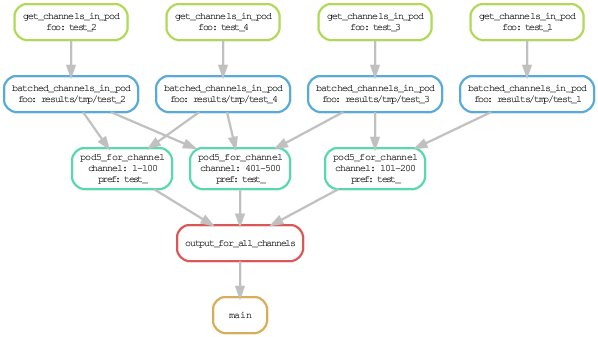
pod5_by_pore
A Snakemake workflow to take the POD5 files produced by an Oxford Nanopore sequencing run and re-batch them by pore (ie. by channel).
This is useful if you want to run duplex basecalling because you can meaningfully run "dorado duplex" on a single (or a subset of) the POD5 files.
Know issues
It is assumed all POD5 input files are from the same sequencing run, but this is not checked.
Version History
master @ 6b1da33 (latest) Created 11th Sep 2024 at 14:31 by Tim Booth
Try using a non-frozen environment.
Frozen
 master
master6b1da33
master @ b2977c5 (earliest) Created 24th May 2024 at 15:28 by Tim Booth
Add a diagram (dot format)
Frozen
 master
masterb2977c5
 Creators and Submitter
Creators and SubmitterCreator
Submitter
License
Activity
Views: 2274 Downloads: 543
Created: 24th May 2024 at 15:28
Last updated: 24th May 2024 at 15:32
Annotated Properties
 Attributions
AttributionsNone
 View on GitHub
View on GitHub https://orcid.org/0000-0003-2470-9519
https://orcid.org/0000-0003-2470-9519