Workflow Type: Galaxy
Open
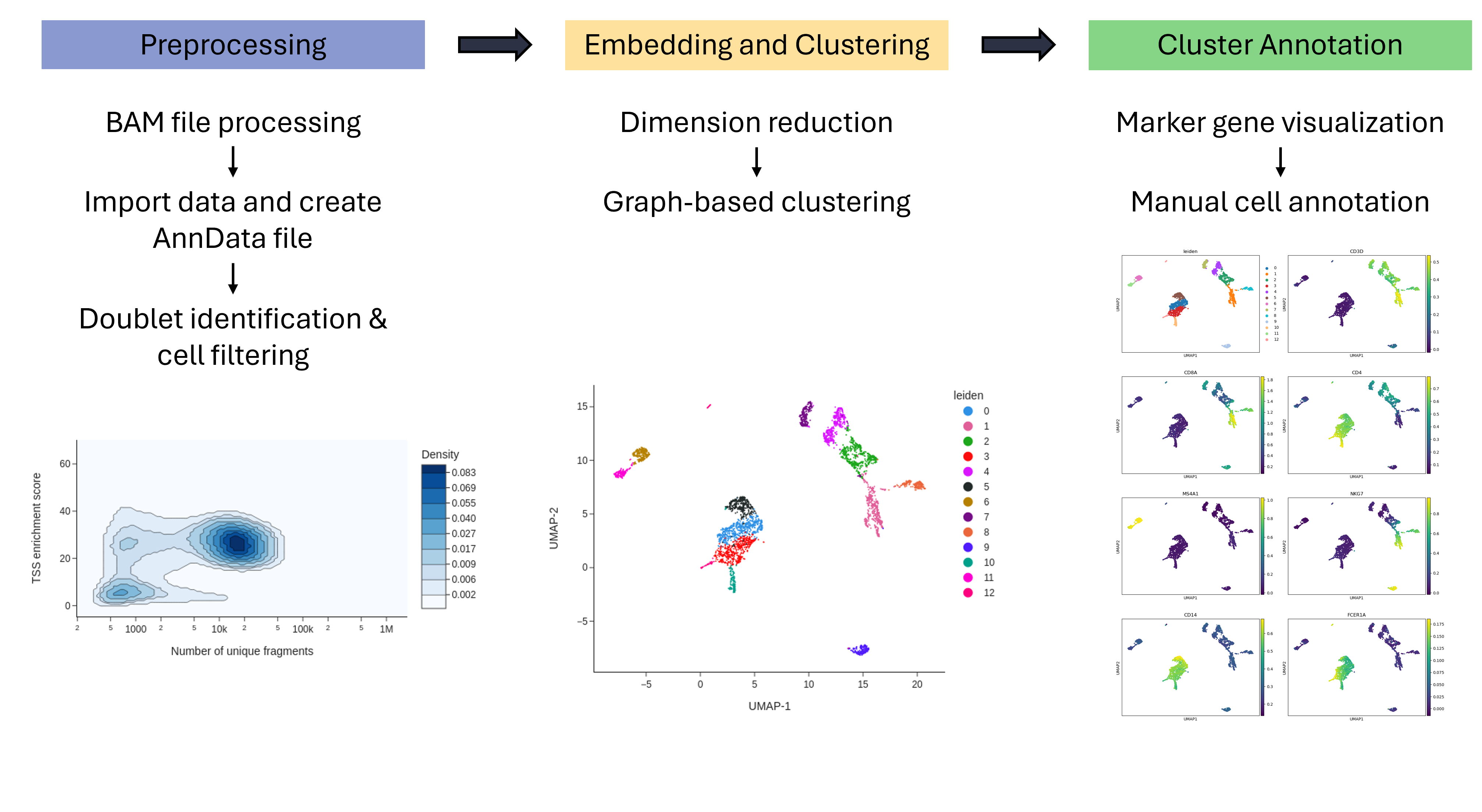
Workflow for Single-cell ATAC-seq standard processing with SnapATAC2. This workflow takes a fragment file as input and performs the standard steps of scATAC-seq analysis: filtering, dimension reduction, embedding and visualization of marker genes with SnapATAC2. Finally, the clusters are manually annotated with the help of marker genes. In an alternative step, the fragment file can also be generated from a BAM file.
- newer Version: Updated SnapATAC2 version from 2.5.3 to 2.6.4
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Bam-file | Bam-file | n/a |
|
| Fragment_file | Fragment_file | n/a |
|
| genes | Keys for annotations of obs/cells or vars/genes | Comma-separated list of obs/cells and vars/genes |
|
| Replace_file | Replace_file | n/a |
|
| chromosome_sizes.tabular | chromosome_sizes.tabular | n/a |
|
| gene_annotation | gene_annotation | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 6 | pp.import_data | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.5.3+galaxy2 |
| 7 | pp.make_fragment_file | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.5.3+galaxy2 |
| 8 | pl.frag_size_distr | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_plotting/snapatac2_plotting/2.5.3+galaxy2 |
| 9 | pl.frag_size_distr_log | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_plotting/snapatac2_plotting/2.5.3+galaxy2 |
| 10 | metrics.tsse | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.5.3+galaxy2 |
| 11 | pp.import_data-sorted_by_barcodes | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.5.3+galaxy2 |
| 12 | pl.tsse | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_plotting/snapatac2_plotting/2.5.3+galaxy2 |
| 13 | pp.filter_cells | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.5.3+galaxy2 |
| 14 | pp.add_tile_matrix | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.5.3+galaxy2 |
| 15 | pp.select_features | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.5.3+galaxy2 |
| 16 | pp.scrublet | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.5.3+galaxy2 |
| 17 | pp.filter_doublets | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.5.3+galaxy2 |
| 18 | tl.spectral | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.5.3+galaxy2 |
| 19 | tl.umap | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.5.3+galaxy2 |
| 20 | pp.knn | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.5.3+galaxy2 |
| 21 | tl.leiden | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.5.3+galaxy2 |
| 22 | pl.umap | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_plotting/snapatac2_plotting/2.5.3+galaxy2 |
| 23 | make_gene_matrix | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.5.3+galaxy2 |
| 24 | scanpy_filter_genes | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.9.6+galaxy3 |
| 25 | Normalize | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_normalize/scanpy_normalize/1.9.6+galaxy3 |
| 26 | pp.log1p | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.9.6+galaxy3 |
| 27 | external.pp.magic | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_normalize/scanpy_normalize/1.9.6+galaxy3 |
| 28 | Copy obsm | toolshed.g2.bx.psu.edu/repos/ebi-gxa/anndata_ops/anndata_ops/1.9.3+galaxy0 |
| 29 | umap_plot_with_scanpy | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.9.6+galaxy3 |
| 30 | Inspect observations | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.3+galaxy0 |
| 31 | Cut leiden from table | Cut1 |
| 32 | Replace leiden | toolshed.g2.bx.psu.edu/repos/bgruening/replace_column_by_key_value_file/replace_column_with_key_value_file/0.2 |
| 33 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.3+galaxy0 |
| 34 | Plot cell types | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.9.6+galaxy3 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| plot frag_size | plot frag_size | n/a |
|
| plot log frag_size | plot log frag_size | n/a |
|
| anndata tsse | anndata tsse | n/a |
|
| anndata | anndata | n/a |
|
| plot tsse | plot tsse | n/a |
|
| anndata filter cells | anndata filter cells | n/a |
|
| anndata tile matrix | anndata tile matrix | n/a |
|
| anndata select features | anndata select features | n/a |
|
| anndata scrublet | anndata scrublet | n/a |
|
| anndata filter doublets | anndata filter doublets | n/a |
|
| anndata spectral | anndata spectral | n/a |
|
| anndata umap | anndata umap | n/a |
|
| anndata knn | anndata knn | n/a |
|
| anndata_leiden_clustering | anndata_leiden_clustering | n/a |
|
| umap_leiden-clusters | umap_leiden-clusters | n/a |
|
| anndata gene matrix | anndata gene matrix | n/a |
|
| anndata filter genes | anndata filter genes | n/a |
|
| anndata normalize | anndata normalize | n/a |
|
| anndata log1p | anndata log1p | n/a |
|
| anndata_magic | anndata_magic | n/a |
|
| anndata_gene-matrix_leiden | anndata_gene-matrix_leiden | n/a |
|
| umap_marker-genes | umap_marker-genes | n/a |
|
| leiden annotation | leiden annotation | n/a |
|
| cell type annotation | cell type annotation | n/a |
|
| anndata_cell_type | anndata_cell_type | n/a |
|
| umap_cell-type | umap_cell-type | n/a |
|
Version History
Version 1 (earliest) Created 17th Jul 2024 at 16:56 by Timon Schlegel
Initial commit
Open
 master
masterc8d3e5f
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Activity
Views: 2541 Downloads: 256 Runs: 3
Created: 17th Jul 2024 at 16:56
Last updated: 2nd Aug 2024 at 19:08
 Tags
Tags Attributions
AttributionsNone
 Run on Galaxy
Run on Galaxy https://orcid.org/0009-0001-3228-105X
https://orcid.org/0009-0001-3228-105X