Workflow Type: Galaxy
Open
Stable
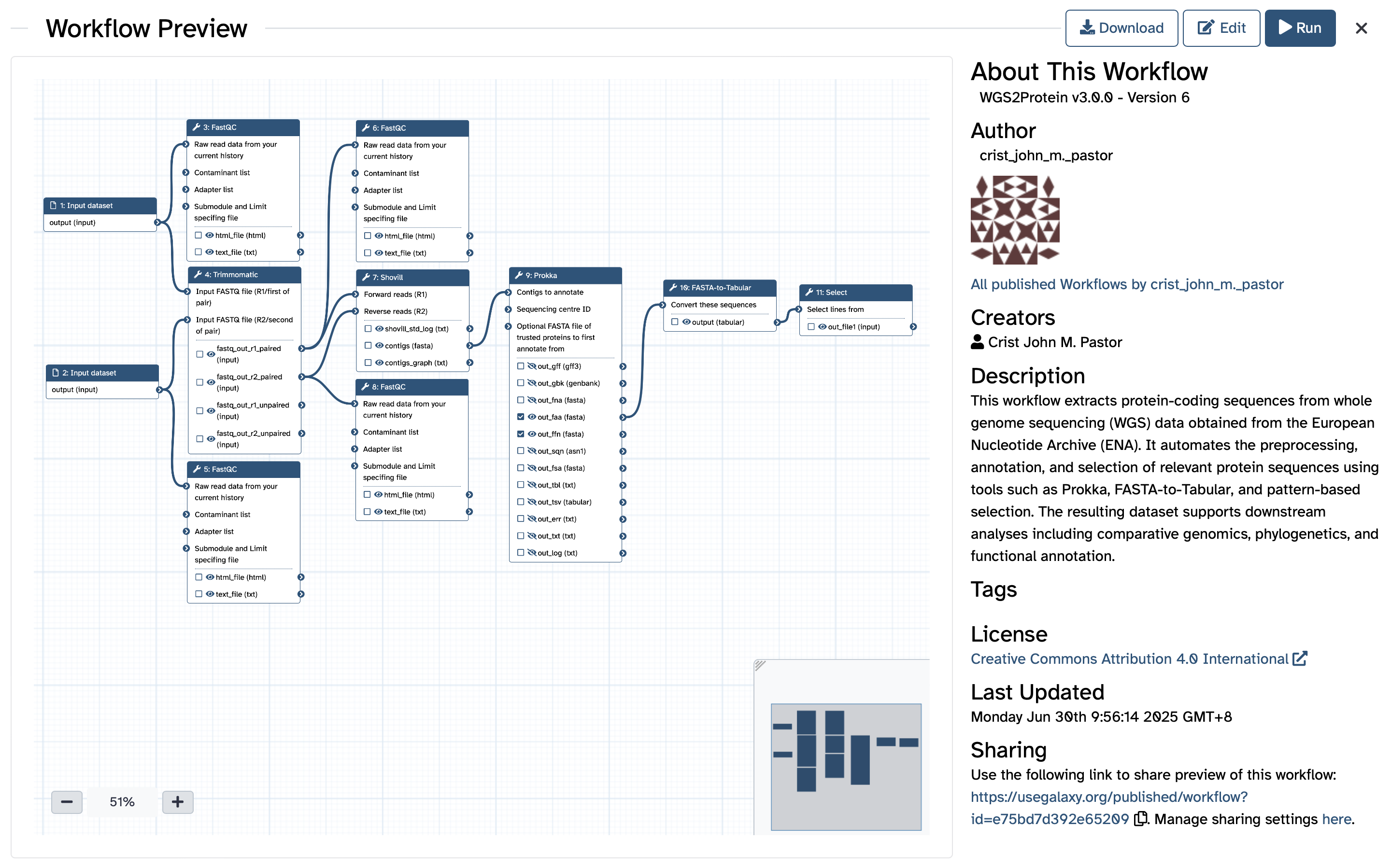
This workflow extracts protein-coding sequences from whole genome sequencing (WGS) data obtained from the European Nucleotide Archive (ENA). It automates the preprocessing, annotation, and selection of relevant protein sequences using tools such as Prokka, FASTA-to-Tabular, and pattern-based selection. The resulting dataset supports downstream analyses including comparative genomics, phylogenetics, and functional annotation.
Steps
| ID | Name | Description |
|---|---|---|
| 2 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 3 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.39+galaxy2 |
| 4 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 5 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 6 | Shovill | toolshed.g2.bx.psu.edu/repos/iuc/shovill/shovill/1.1.0+galaxy2 |
| 7 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 8 | Prokka | toolshed.g2.bx.psu.edu/repos/crs4/prokka/prokka/1.14.6+galaxy1 |
| 9 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.1 |
| 10 | Select | Grep1 |
Version History
Version 1 (earliest) Created 30th Jun 2025 at 10:26 by Crist John Pastor
Initial commit
Open
 master
master8a0242c
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Tools
Activity
Views: 13 Downloads: 4 Runs: 1
Created: 30th Jun 2025 at 10:26
Annotated Properties
Topic annotations
Drug discovery, Immunology, Drug development, Immunoproteins and antigens, Immunoinformatics, Biochemistry, Data mining, Proteins
Operation annotations
 Tags
TagsThis item has not yet been tagged.
 Attributions
AttributionsNone
 Run on Galaxy
Run on Galaxy https://orcid.org/0000-0001-5796-3068
https://orcid.org/0000-0001-5796-3068