Workflow Type: Galaxy
Open
Frozen
Stable
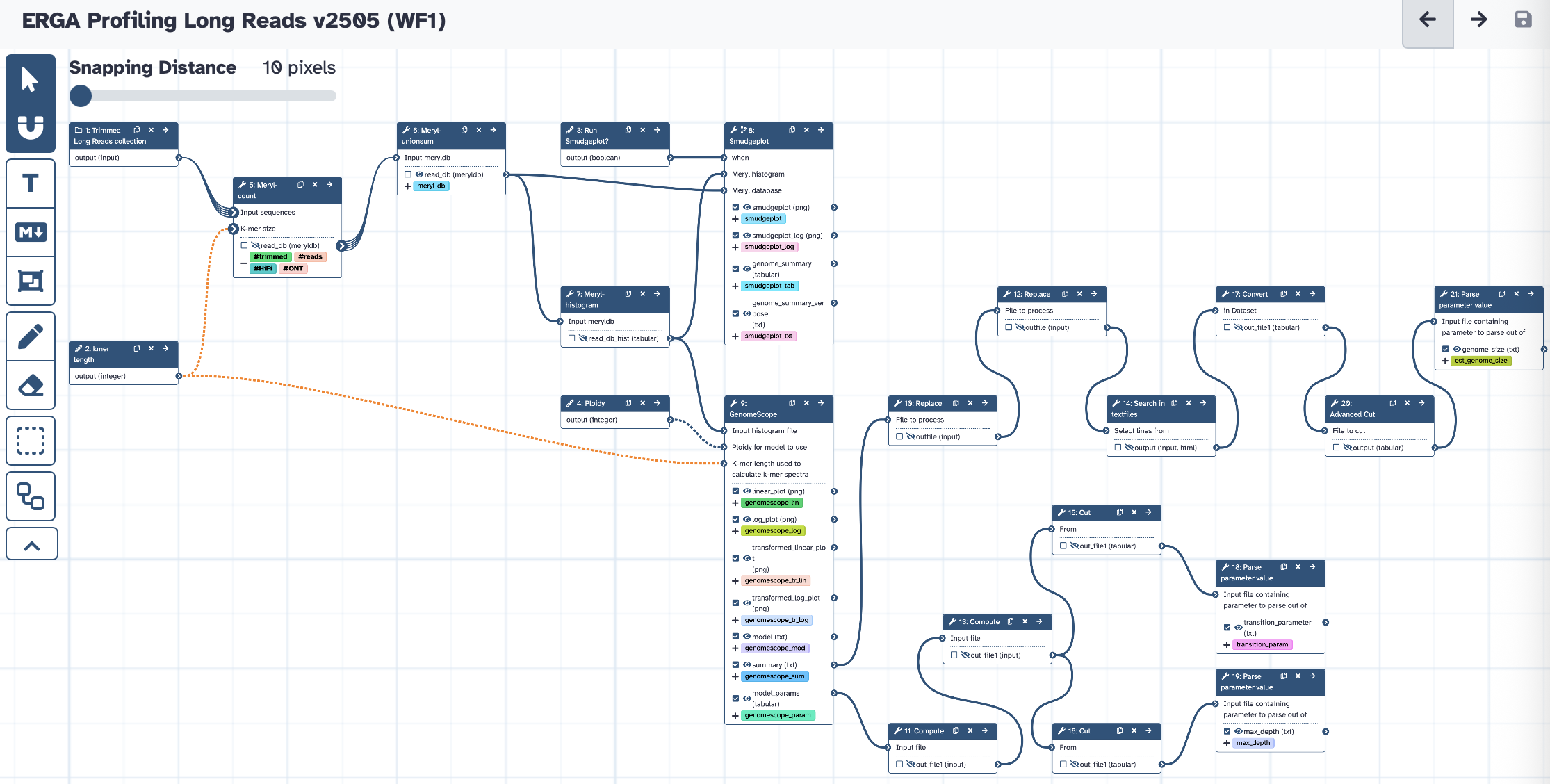
The workflow takes a (trimmed) Long reads collection, runs Meryl to create a K-mer database, Genomescope2 to estimate genome properties and Smudgeplot to estimate ploidy (optional). The main results are K-mer database and genome profiling plots, tables, and values useful for downstream analysis. Default K-mer length and ploidy for Genomescope are 31 and 2, respectively.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Ploidy | Ploidy | Default ploidy: 2 |
|
| Run Smudgeplot? | Run Smudgeplot? | n/a |
|
| Trimmed Long Reads collection | Trimmed Long Reads collection | n/a |
|
| kmer length | kmer length | Kmer length for calculating kmer spectra |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Meryl-count | toolshed.g2.bx.psu.edu/repos/iuc/meryl/meryl/1.3+galaxy6 |
| 5 | Meryl-unionsum | toolshed.g2.bx.psu.edu/repos/iuc/meryl/meryl/1.3+galaxy6 |
| 6 | Meryl-histogram | toolshed.g2.bx.psu.edu/repos/iuc/meryl/meryl/1.3+galaxy6 |
| 7 | Smudgeplot | toolshed.g2.bx.psu.edu/repos/galaxy-australia/smudgeplot/smudgeplot/0.2.5+galaxy3 |
| 8 | GenomeScope | toolshed.g2.bx.psu.edu/repos/iuc/genomescope/genomescope/2.0.1+galaxy0 |
| 9 | Replace | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_find_and_replace/1.1.4 |
| 10 | Compute | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.0 |
| 11 | Replace | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_find_and_replace/1.1.4 |
| 12 | Compute | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.0 |
| 13 | Search in textfiles | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/1.1.1 |
| 14 | Cut | Cut1 |
| 15 | Cut | Cut1 |
| 16 | Convert | Convert characters1 |
| 17 | Parse parameter value | param_value_from_file |
| 18 | Parse parameter value | param_value_from_file |
| 19 | Advanced Cut | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/1.1.0 |
| 20 | Parse parameter value | param_value_from_file |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
| _anonymous_output_2 | _anonymous_output_2 | n/a |
|
| _anonymous_output_3 | _anonymous_output_3 | n/a |
|
| _anonymous_output_4 | _anonymous_output_4 | n/a |
|
| _anonymous_output_5 | _anonymous_output_5 | n/a |
|
| _anonymous_output_6 | _anonymous_output_6 | n/a |
|
| _anonymous_output_7 | _anonymous_output_7 | n/a |
|
| _anonymous_output_8 | _anonymous_output_8 | n/a |
|
| _anonymous_output_9 | _anonymous_output_9 | n/a |
|
| _anonymous_output_10 | _anonymous_output_10 | n/a |
|
| _anonymous_output_11 | _anonymous_output_11 | n/a |
|
| transition_parameter | transition_parameter | n/a |
|
| max_depth | max_depth | n/a |
|
| genome_size | genome_size | n/a |
|
Version History
Version 2 (latest) Created 18th Sep 2024 at 14:27 by Diego De Panis
No revision comments
Open
 master
master8e191f8
Version 1 (earliest) Created 6th Oct 2023 at 14:25 by Diego De Panis
Initial commit
Frozen
 Version-1
Version-16911bb8
 Creators and Submitter
Creators and SubmitterCreator
Additional credit
ERGA
Submitter
Discussion Channel
Tools
License
Activity
Views: 5793 Downloads: 772 Runs: 75
Created: 6th Oct 2023 at 14:25
Last updated: 24th Jun 2025 at 16:41
Annotated Properties
 Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-3679-9585
https://orcid.org/0000-0002-3679-9585