Workflow Type: Galaxy
Open
Stable
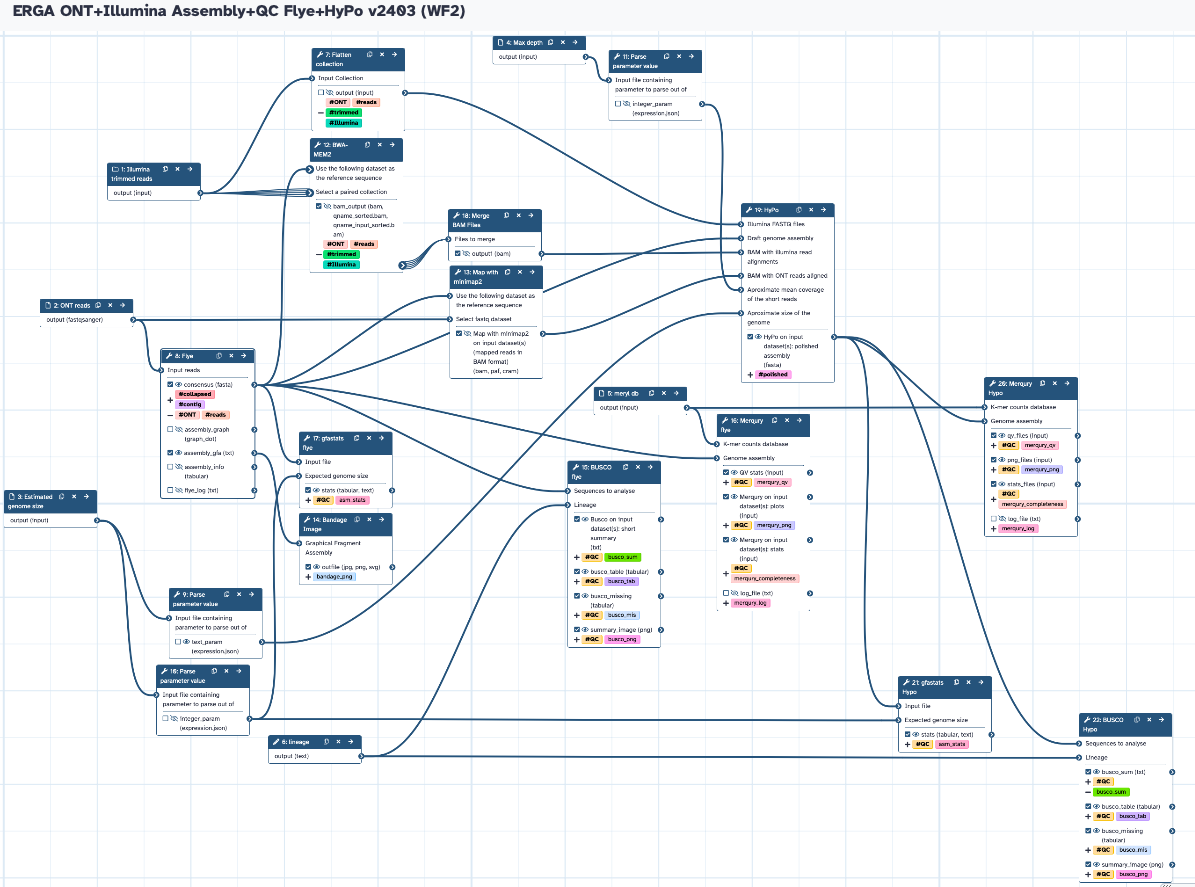
The workflow takes raw ONT reads and trimmed Illumina WGS paired reads collections, and the estimated genome size and Max depth (both calculated from WF1) to run Flye and subsequently polish the assembly with HyPo. It produces collapsed assemblies (unpolished and polished) and runs all the QC analyses (gfastats, BUSCO, and Merqury).
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Estimated genome size | Estimated genome size | n/a |
|
| Illumina trimmed reads | Illumina trimmed reads | n/a |
|
| Max depth | Max depth | n/a |
|
| ONT reads | ONT reads | fastq file with ONT reads |
|
| lineage | lineage | lineage for BUSCO, e.g.: arthropoda_odb10, vertebrata_odb10, mammalia_odb10... |
|
| meryl db | meryl db | import meryldb to run merqury on the assemblies obtained |
|
Steps
| ID | Name | Description |
|---|---|---|
| 6 | Flatten collection | __FLATTEN__ |
| 7 | Flye | toolshed.g2.bx.psu.edu/repos/bgruening/flye/flye/2.9.1+galaxy0 |
| 8 | Parse parameter value | param_value_from_file |
| 9 | Parse parameter value | param_value_from_file |
| 10 | Parse parameter value | param_value_from_file |
| 11 | BWA-MEM2 | toolshed.g2.bx.psu.edu/repos/iuc/bwa_mem2/bwa_mem2/2.2.1+galaxy0 |
| 12 | Map with minimap2 | toolshed.g2.bx.psu.edu/repos/iuc/minimap2/minimap2/2.26+galaxy0 |
| 13 | Bandage Image | toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_image/2022.09+galaxy4 |
| 14 | BUSCO flye | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.4.6+galaxy0 |
| 15 | Merqury flye | toolshed.g2.bx.psu.edu/repos/iuc/merqury/merqury/1.3+galaxy2 |
| 16 | gfastats flye | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.6+galaxy0 |
| 17 | Merge BAM Files | toolshed.g2.bx.psu.edu/repos/devteam/sam_merge/sam_merge2/1.2.0 |
| 18 | HyPo | toolshed.g2.bx.psu.edu/repos/iuc/hypo/hypo/1.0.3+galaxy0 |
| 19 | Merqury Hypo | merqury on flye assembly polished with 1 round of hypo toolshed.g2.bx.psu.edu/repos/iuc/merqury/merqury/1.3+galaxy2 |
| 20 | gfastats Hypo | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.6+galaxy0 |
| 21 | BUSCO Hypo | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.4.6+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| consensus | consensus | n/a |
|
| assembly_gfa | assembly_gfa | n/a |
|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
| Map with minimap2 on input dataset(s) (mapped reads in BAM format) | Map with minimap2 on input dataset(s) (mapped reads in BAM format) | n/a |
|
| _anonymous_output_2 | _anonymous_output_2 | n/a |
|
| Busco on input dataset(s): short summary | Busco on input dataset(s): short summary | n/a |
|
| _anonymous_output_3 | _anonymous_output_3 | n/a |
|
| _anonymous_output_4 | _anonymous_output_4 | n/a |
|
| _anonymous_output_5 | _anonymous_output_5 | n/a |
|
| Merqury on input dataset(s): plots | Merqury on input dataset(s): plots | n/a |
|
| QV stats | QV stats | n/a |
|
| Merqury on input dataset(s): stats | Merqury on input dataset(s): stats | n/a |
|
| _anonymous_output_6 | _anonymous_output_6 | n/a |
|
| output1 | output1 | n/a |
|
| HyPo on input dataset(s): polished assembly | HyPo on input dataset(s): polished assembly | n/a |
|
| _anonymous_output_7 | _anonymous_output_7 | n/a |
|
| _anonymous_output_8 | _anonymous_output_8 | n/a |
|
| _anonymous_output_9 | _anonymous_output_9 | n/a |
|
| _anonymous_output_10 | _anonymous_output_10 | n/a |
|
| _anonymous_output_11 | _anonymous_output_11 | n/a |
|
| _anonymous_output_12 | _anonymous_output_12 | n/a |
|
| _anonymous_output_13 | _anonymous_output_13 | n/a |
|
| _anonymous_output_14 | _anonymous_output_14 | n/a |
|
Version History
Version 1 (earliest) Created 11th Mar 2024 at 12:41 by Diego De Panis
Initial commit
Open
 master
mastera0cb403
 Creators and Submitter
Creators and SubmitterCreator
Additional credit
ERGA
Submitter
Discussion Channel
License
Activity
Views: 3531 Downloads: 387 Runs: 5
Created: 11th Mar 2024 at 12:41
Annotated Properties
 Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-3679-9585
https://orcid.org/0000-0002-3679-9585