Workflow Type: Galaxy
Open
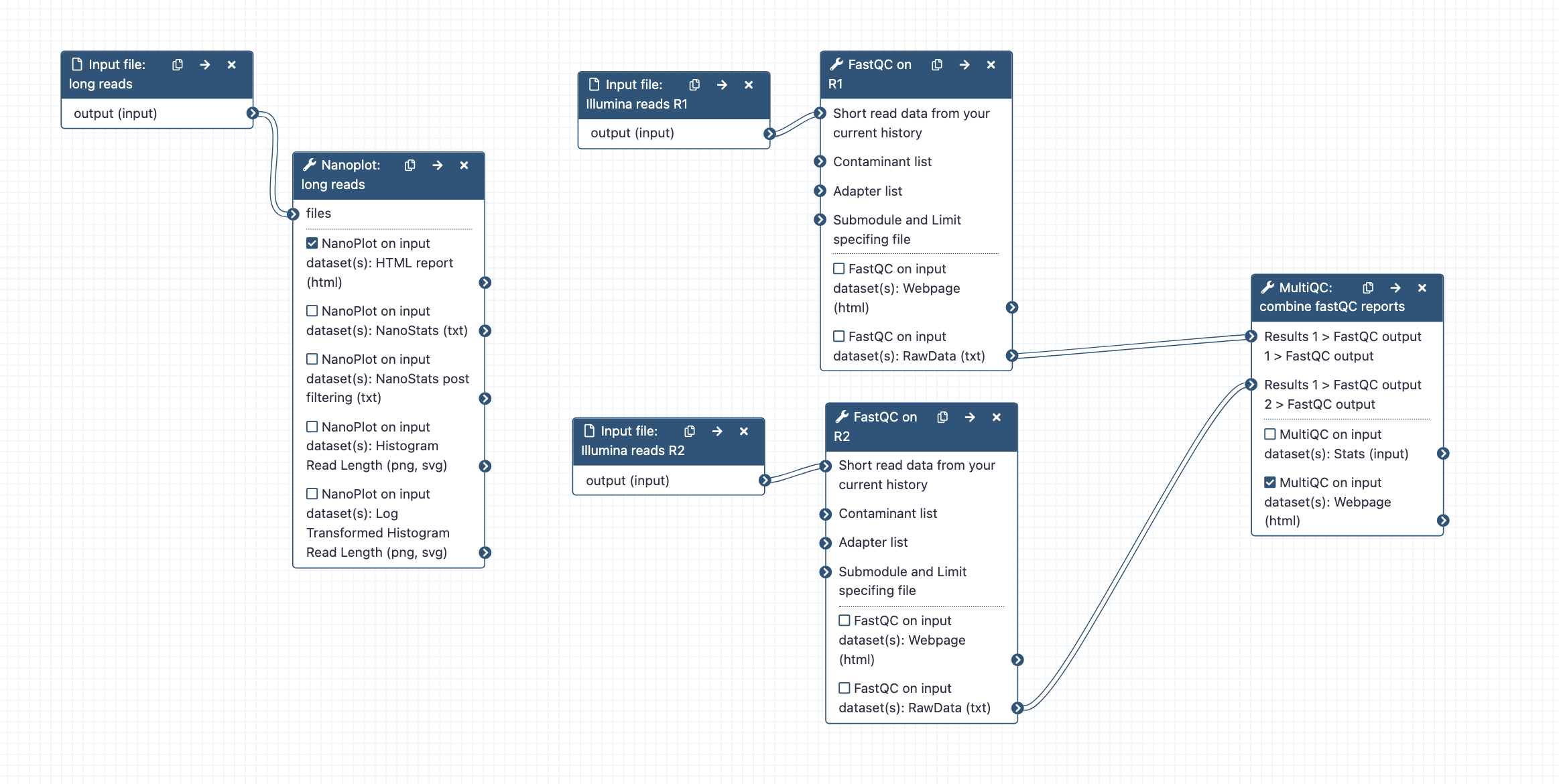
Data QC step, can run alone or as part of a combined workflow for large genome assembly.
- What it does: Reports statistics from sequencing reads.
- Inputs: long reads (fastq.gz format), short reads (R1 and R2) (fastq.gz format).
- Outputs: For long reads: a nanoplot report (the HTML report summarizes all the information). For short reads: a MultiQC report.
- Tools used: Nanoplot, FastQC, MultiQC.
- Input parameters: None required.
- Workflow steps: Long reads are analysed by Nanoplot; Short reads (R1 and R2) are analysed by FastQC; the resulting reports are processed by MultiQC.
- Options: see the tool settings options at runtime and change as required. Alternative tool option: fastp
Infrastructure_deployment_metadata: Galaxy Australia (Galaxy)
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Input file: Illumina reads R1 | Input file: Illumina reads R1 | n/a |
|
| Input file: Illumina reads R2 | Input file: Illumina reads R2 | n/a |
|
| Input file: long reads | Input file: long reads | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Nanoplot: long reads | toolshed.g2.bx.psu.edu/repos/iuc/nanoplot/nanoplot/1.28.2+galaxy1 |
| 4 | FastQC on R1 | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 5 | FastQC on R2 | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 6 | MultiQC: combine fastQC reports | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.9+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| NanoPlot on input dataset(s): HTML report | NanoPlot on input dataset(s): HTML report | n/a |
|
| MultiQC on input dataset(s): Webpage | MultiQC on input dataset(s): Webpage | n/a |
|
Version History
Version 1 (earliest) Created 8th Nov 2021 at 04:34 by Anna Syme
Added/updated 2 files
Open
 master
masterc962e98
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Citation
Syme, A. (2021). Data QC. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.222.1
License
Activity
Views: 6217 Downloads: 525 Runs: 1
Created: 8th Nov 2021 at 04:34
Last updated: 9th Nov 2021 at 01:09
Annotated Properties
 Attributions
AttributionsNone
 Collections
Collections Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673