Workflow Type: Galaxy
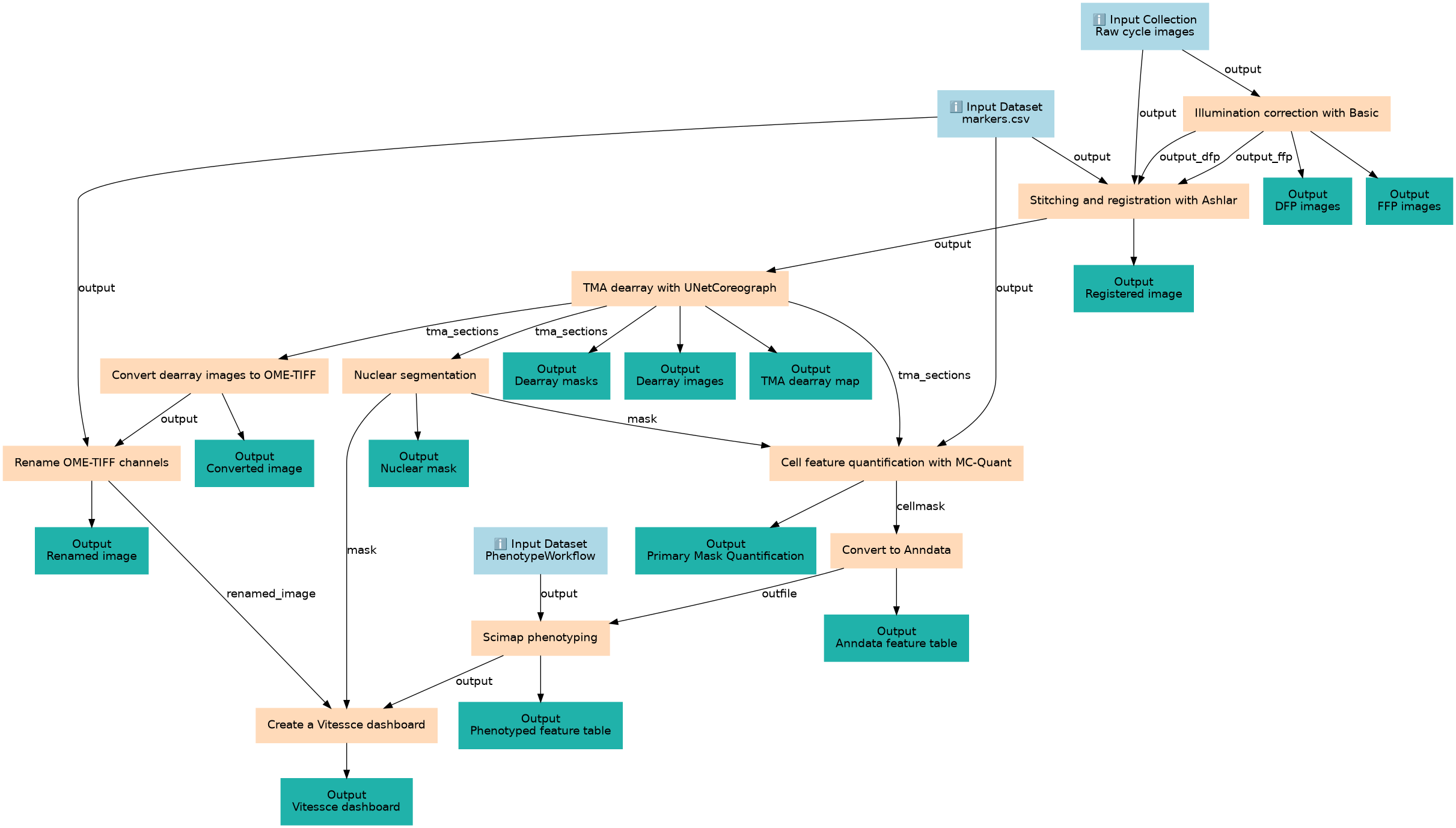
End-to-End Tissue Microarray Analysis with Galaxy-ME
Associated Tutorial
This workflows is part of the tutorial End-to-End Tissue Microarray Image Analysis with Galaxy-ME, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): Cameron Watson
Tutorial Author(s): Cameron Watson, Allison Creason
Tutorial Contributor(s): Leonid Kostrykin, Beatriz Serrano-Solano, Helena Rasche, Saskia Hiltemann, Cameron Watson
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| PhenotypeWorkflow | #main/PhenotypeWorkflow | Scimap formatted phenotype workflow |
|
| Raw cycle images | #main/Raw cycle images | Raw TIFF images (CZI, TIFF) in an round-ordered list |
|
| markers.csv | #main/markers.csv | CSV file containing marker names |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Illumination correction with Basic | Illumination correction toolshed.g2.bx.psu.edu/repos/perssond/basic_illumination/basic_illumination/1.1.1+galaxy2 |
| 4 | Stitching and registration with Ashlar | Stitching and registration toolshed.g2.bx.psu.edu/repos/perssond/ashlar/ashlar/1.18.0+galaxy1 |
| 5 | TMA dearray with UNetCoreograph | TMA dearray toolshed.g2.bx.psu.edu/repos/perssond/coreograph/unet_coreograph/2.2.8+galaxy1 |
| 6 | Nuclear segmentation | Nuclear segmentation toolshed.g2.bx.psu.edu/repos/goeckslab/mesmer/mesmer/0.12.3+galaxy3 |
| 7 | Convert dearray images to OME-TIFF | Convert to OME-TIFF toolshed.g2.bx.psu.edu/repos/imgteam/bfconvert/ip_convertimage/6.7.0+galaxy0 |
| 8 | Cell feature quantification with MC-Quant | Quantify cell features toolshed.g2.bx.psu.edu/repos/perssond/quantification/quantification/1.6.0+galaxy0 |
| 9 | Rename OME-TIFF channels | Rename OME-TIFF channels toolshed.g2.bx.psu.edu/repos/goeckslab/rename_tiff_channels/rename_tiff_channels/0.0.2+galaxy1 |
| 10 | Convert to Anndata | Convert to Anndata toolshed.g2.bx.psu.edu/repos/goeckslab/scimap_mcmicro_to_anndata/scimap_mcmicro_to_anndata/2.1.0+galaxy2 |
| 11 | Scimap phenotyping | Scimap phenotyping toolshed.g2.bx.psu.edu/repos/goeckslab/scimap_phenotyping/scimap_phenotyping/2.1.0+galaxy2 |
| 12 | Create a Vitessce dashboard | Create a Vitessce dashboard toolshed.g2.bx.psu.edu/repos/goeckslab/vitessce_spatial/vitessce_spatial/3.5.1+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Anndata feature table | #main/Anndata feature table | n/a |
|
| Converted image | #main/Converted image | n/a |
|
| DFP images | #main/DFP images | n/a |
|
| Dearray images | #main/Dearray images | n/a |
|
| Dearray masks | #main/Dearray masks | n/a |
|
| FFP images | #main/FFP images | n/a |
|
| Nuclear mask | #main/Nuclear mask | n/a |
|
| Phenotyped feature table | #main/Phenotyped feature table | n/a |
|
| Primary Mask Quantification | #main/Primary Mask Quantification | n/a |
|
| Registered image | #main/Registered image | n/a |
|
| Renamed image | #main/Renamed image | n/a |
|
| TMA dearray map | #main/TMA dearray map | n/a |
|
| Vitessce dashboard | #main/Vitessce dashboard | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 228 Downloads: 28 Runs: 0
Created: 2nd Jun 2025 at 10:56
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master