Workflow Type: Galaxy
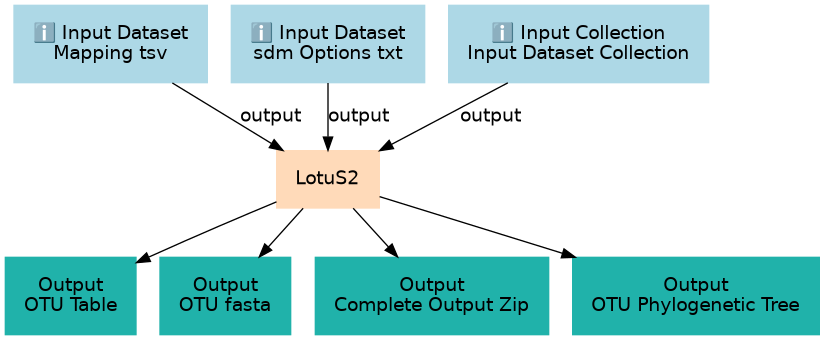
Workflow for running LotuS2 tool on fungal ITS paired-end sequencing data, to identify the fungi present in the samples
Associated Tutorial
This workflows is part of the tutorial Identifying Mycorrhizal Fungi from ITS2 sequencing using LotuS2, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Society for the Protection of Underground Networks, Sujai Kumar, Bethan Manley
Tutorial Author(s): Sujai Kumar
Tutorial Contributor(s): Bethan Manley, Paul Zierep, Helena Rasche, Saskia Hiltemann, Sujai Kumar
Funder(s): Society for the Protection of Underground Networks
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Input Dataset Collection | #main/Input Dataset Collection | List of paired fastq files needed to run LotuS2. These files MUST match the files in the Mapping File |
|
| Mapping tsv | #main/Mapping tsv | Mapping file needed by LotuS2 to link sample metadata to a pair of fastq files |
|
| sdm Options txt | #main/sdm Options txt | SDM (sequence demultiplexer tool) options needed for fungal ITS illumina paired-end sequencing |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | LotuS2 | LotuS2 creates a large output folder which is zipped up and provided as one of the outputs. Some of the individual files within this output folder are also exposed as outputs by the Galaxy LotuS2 tool so that they can be used in subsequent downstream analyses. toolshed.g2.bx.psu.edu/repos/earlhaminst/lotus2/lotus2/2.32+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Complete Output Zip | #main/Complete Output Zip | n/a |
|
| OTU Phylogenetic Tree | #main/OTU Phylogenetic Tree | n/a |
|
| OTU Table | #main/OTU Table | n/a |
|
| OTU fasta | #main/OTU fasta | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tool
License
Activity
Views: 231 Downloads: 32 Runs: 0
Created: 2nd Jun 2025 at 10:54
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master