Workflow Type: Galaxy
Frozen
This is part of a series of workflows to annotate a genome, tagged with TSI-annotation.
These workflows are based on command-line code by Luke Silver, converted into Galaxy Australia workflows.
The workflows can be run in this order:
- Repeat masking
- RNAseq QC and read trimming
- Find transcripts
- Combine transcripts
- Extract transcripts
- Convert formats
- Fgenesh annotation
About this workflow:
- Repeat this workflow separately for datasets from different tissues.
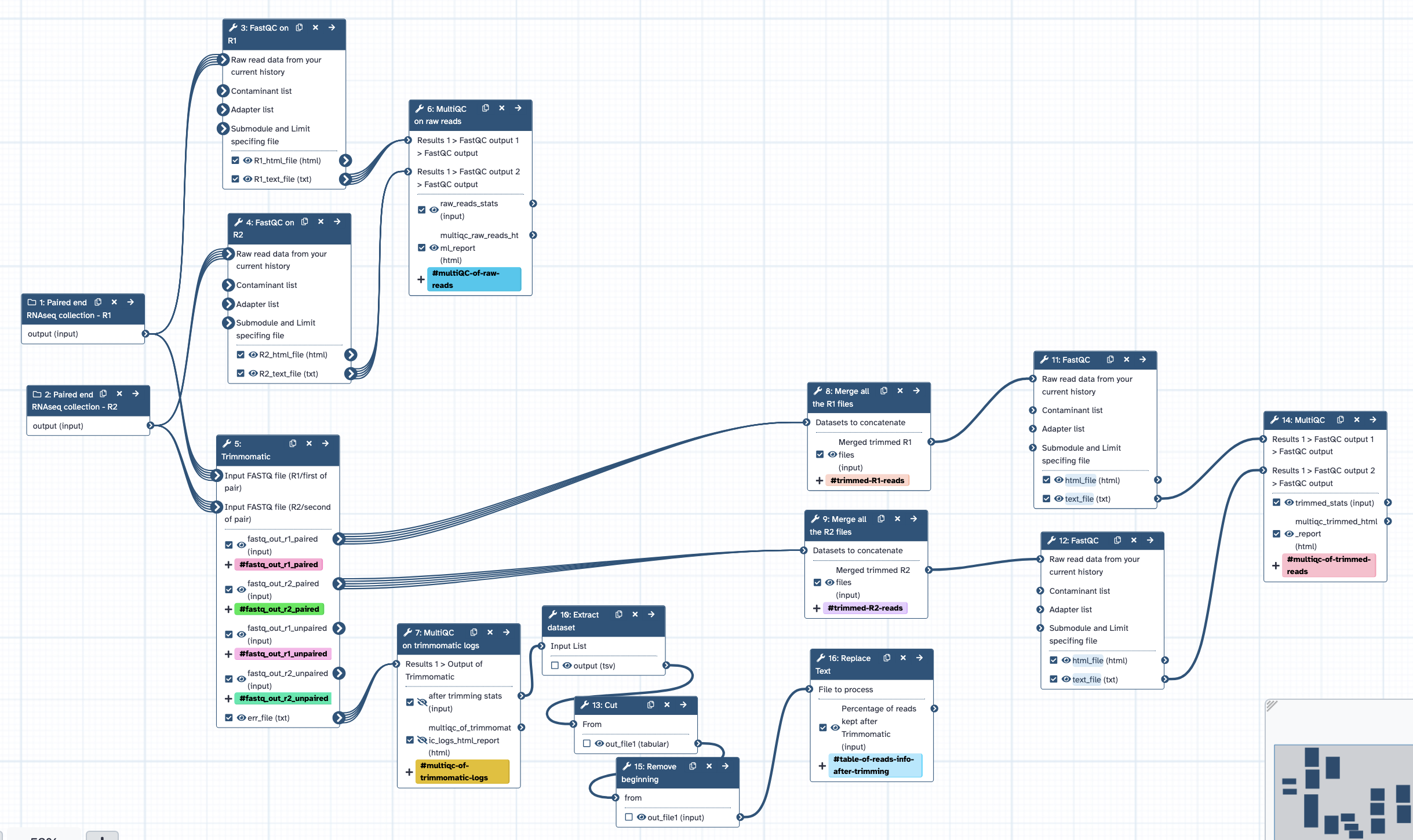
- Inputs = collections of R1 files, and R2 files (all from a single tissue type).
- Runs FastQC with default settings, separately for raw reads R1 and R2 collections; all output to MultiQC.
- Runs Trimmomatic with initial ILLUMINACLIP step (using standard adapter sequence for TruSeq3 paired-ended), uses settings SLIDINGWINDOW:4:5 LEADING:5 TRAILING:5 MINLEN:25, retain paired (not unpaired) outputs. User can modify at runtime.
- Runs FastQC with default settings, separately for trimmed R1 and R2 collections; all output to MultiQC.
- From Trimmomatic output: concatenate all R1 reads; concatenate all R2 reads.
- Outputs = trimmed merged R1 file, trimmed merged R2 file.
- Log files from Trimmomatic to MultiQC, to summarise trimming results.
- Note: a known bug with MultiQC html output is that plot is labelled as "R1" reads, when it actually contains information from both R1 and R2 read sets - this is under investigation (and is due to a Trimmomatic output file labelling issue).
- MultiQC results table formatted to show % of reads retained after trimming, table included in workflow report.
- Note: a known bug is that sometimes the workflow report text resets to default text. To restore, look for an earlier workflow version with correct workflow report text, and copy and paste report text into current version.
Steps
| ID | Name | Description |
|---|---|---|
| 2 | FastQC on R1 | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 3 | FastQC on R2 | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 4 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.36.6 |
| 5 | MultiQC on raw reads | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 6 | MultiQC on trimmomatic logs | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 7 | Merge all the R1 files | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cat/0.1.1 |
| 8 | Merge all the R2 files | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cat/0.1.1 |
| 9 | Extract dataset | __EXTRACT_DATASET__ |
| 10 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 11 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 12 | Cut | Cut1 |
| 13 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 14 | Remove beginning | Remove beginning1 |
| 15 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3 |
Version History
Version 1 (earliest) Created 8th May 2024 at 07:39 by Anna Syme
Initial commit
Frozen
 Version-1
Version-10aa7186
 Creators and Submitter
Creators and SubmitterCreators
Submitter
Tools
Citation
Silver, L., & Syme, A. (2024). QC and trimming of RNAseq reads - TSI. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.876.1
Activity
Views: 2520 Downloads: 244 Runs: 4
Created: 8th May 2024 at 07:39
Last updated: 9th May 2024 at 05:02
 Tags
Tags Attributions
AttributionsNone
 Collections
Collections Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673