Workflow Type: Galaxy
Open
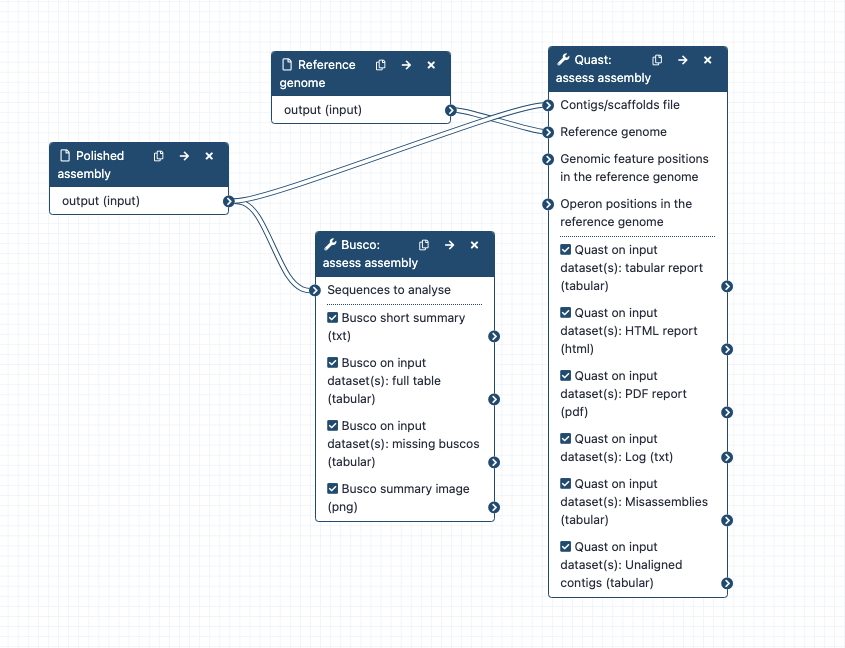
Assess genome quality; can run alone or as part of a combined workflow for large genome assembly.
- What it does: Assesses the quality of the genome assembly: generate some statistics and determine if expected genes are present; align contigs to a reference genome.
- Inputs: polished assembly; reference_genome.fasta (e.g. of a closely-related species, if available).
- Outputs: Busco table of genes found; Quast HTML report, and link to Icarus contigs browser, showing contigs aligned to a reference genome
- Tools used: Busco, Quast
- Input parameters: None required
Workflow steps:
Polished assembly => Busco
- First: predict genes in the assembly: using Metaeuk
- Second: compare the set of predicted genes to the set of expected genes in a particular lineage. Default setting for lineage: Eukaryota
Polished assembly and a reference genome => Quast
- Contigs/scaffolds file: polished assembly
- Type of assembly: Genome
- Use a reference genome: Yes
- Reference genome: Arabidopsis genome
- Is the genome large (> 100Mbp)? Yes.
- All other settings as defaults, except second last setting: Distinguish contigs with more than 50% unaligned bases as a separate group of contigs?: change to No
Options
Gene prediction:
- Change tool used by Busco to predict genes in the assembly: instead of Metaeuk, use Augustus.
- To do this: select: Use Augustus; Use another predefined species model; then choose from the drop down list.
- Select from a database of trained species models. list here: https://github.com/Gaius-Augustus/Augustus/tree/master/config/species
- Note: if using Augustus: it may fail if the input assembly is too small (e.g. a test-size data assembly). It can't do the training part properly.
Compare genes found to other lineage:
- Busco has databases of lineages and their expected genes. Option to change lineage.
- Not all lineages are available - there is a mix of broader and narrower lineages. - list of lineages here: https://busco.ezlab.org/list_of_lineages.html.
- To see the groups in taxonomic hierarchies: Eukaryotes: https://busco.ezlab.org/frames/euka.htm
- For example, if you have a plant species from Fabales, you could set that as the lineage.
- The narrower the taxonomic group, the more total genes are expected.
Infrastructure_deployment_metadata: Galaxy Australia (Galaxy)
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Polished assembly | Polished assembly | n/a |
|
| Reference genome | Reference genome | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Busco: assess assembly | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.2.2+galaxy0 |
| 3 | Quast: assess assembly | toolshed.g2.bx.psu.edu/repos/iuc/quast/quast/5.0.2+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
| Busco summary image | Busco summary image | n/a |
|
| _anonymous_output_2 | _anonymous_output_2 | n/a |
|
| Busco short summary | Busco short summary | n/a |
|
| Quast on input dataset(s): Log | Quast on input dataset(s): Log | n/a |
|
| _anonymous_output_3 | _anonymous_output_3 | n/a |
|
| Quast on input dataset(s): PDF report | Quast on input dataset(s): PDF report | n/a |
|
| Quast on input dataset(s): tabular report | Quast on input dataset(s): tabular report | n/a |
|
| Quast on input dataset(s): HTML report | Quast on input dataset(s): HTML report | n/a |
|
| _anonymous_output_4 | _anonymous_output_4 | n/a |
|
Version History
Version 1 (earliest) Created 8th Nov 2021 at 06:03 by Anna Syme
Added/updated 2 files
Open
 master
mastera760082
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Citation
Syme, A. (2021). Assess genome quality. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.229.1
License
Activity
Views: 6263 Downloads: 480 Runs: 0
Created: 8th Nov 2021 at 06:03
Last updated: 9th Nov 2021 at 01:12
Annotated Properties
Topic annotations
 Attributions
AttributionsNone
 Collections
Collections Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673