Workflow Type: Galaxy
Open
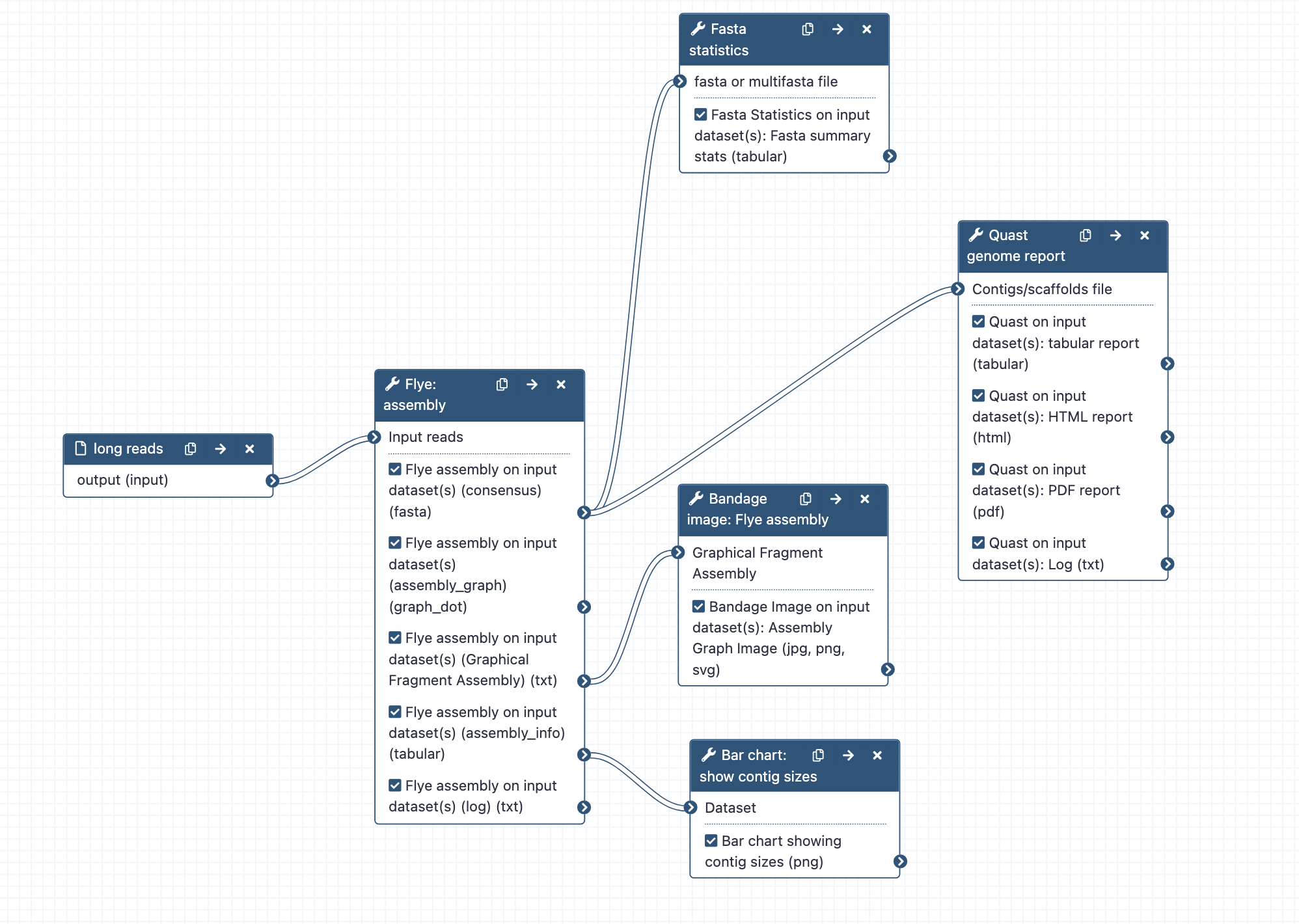
Assembly with Flye; can run alone or as part of a combined workflow for large genome assembly.
- What it does: Assembles long reads with the tool Flye
- Inputs: long reads (may be raw, or filtered, and/or corrected); fastq.gz format
- Outputs: Flye assembly fasta; Fasta stats on assembly.fasta; Assembly graph image from Bandage; Bar chart of contig sizes; Quast reports of genome assembly
- Tools used: Flye, Fasta statistics, Bandage, Bar chart, Quast
- Input parameters: None required, but recommend setting assembly mode to match input sequence type
Workflow steps:
- Long reads are assembled with Flye, using default tool settings. Note: the default setting for read type ("mode") is nanopore raw. Change this at runtime if required.
- Statistics are computed from the assembly.fasta file output, using Fasta Statistics and Quast (is genome large: Yes; distinguish contigs with more that 50% unaligned bases: no)
- The graphical fragment assembly file is visualized with the tool Bandage.
- Assembly information sent to bar chart to visualize contig sizes
Options
- See other Flye options.
- Use a different assembler (in a different workflow).
- Bandage image options - change size (max size is 32767), labels - add (e.g. node lengths). You can also install Bandage on your own computer and donwload the "graphical fragment assembly" file to view in greater detail.
Infrastructure_deployment_metadata: Galaxy Australia (Galaxy)
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| long reads | long reads | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | Flye: assembly | toolshed.g2.bx.psu.edu/repos/bgruening/flye/flye/2.8.2+galaxy0 |
| 2 | Fasta statistics | toolshed.g2.bx.psu.edu/repos/iuc/fasta_stats/fasta-stats/1.0.3 |
| 3 | Bandage image: Flye assembly | toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_image/0.8.1+galaxy3 |
| 4 | Quast genome report | toolshed.g2.bx.psu.edu/repos/iuc/quast/quast/5.0.2+galaxy1 |
| 5 | Bar chart: show contig sizes | barchart_gnuplot |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Flye assembly on input dataset(s) (consensus) | Flye assembly on input dataset(s) (consensus) | n/a |
|
| Flye assembly on input dataset(s) (assembly_graph) | Flye assembly on input dataset(s) (assembly_graph) | n/a |
|
| Flye assembly on input dataset(s) (Graphical Fragment Assembly) | Flye assembly on input dataset(s) (Graphical Fragment Assembly) | n/a |
|
| Flye assembly on input dataset(s) (assembly_info) | Flye assembly on input dataset(s) (assembly_info) | n/a |
|
| Flye assembly on input dataset(s) (log) | Flye assembly on input dataset(s) (log) | n/a |
|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
| Bandage Image on input dataset(s): Assembly Graph Image | Bandage Image on input dataset(s): Assembly Graph Image | n/a |
|
| Quast on input dataset(s): Log | Quast on input dataset(s): Log | n/a |
|
| Quast on input dataset(s): PDF report | Quast on input dataset(s): PDF report | n/a |
|
| Quast on input dataset(s): tabular report | Quast on input dataset(s): tabular report | n/a |
|
| Quast on input dataset(s): HTML report | Quast on input dataset(s): HTML report | n/a |
|
| Bar chart showing contig sizes | Bar chart showing contig sizes | n/a |
|
Version History
Version 1 (earliest) Created 8th Nov 2021 at 05:07 by Anna Syme
Added/updated 2 files
Open
 master
master9541ba8
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Citation
Syme, A. (2021). Assembly with Flye. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.225.1
License
Activity
Views: 7952 Downloads: 529 Runs: 1
Created: 8th Nov 2021 at 05:07
Last updated: 9th Nov 2021 at 01:11
Annotated Properties
 Attributions
AttributionsNone
 Collections
Collections Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673