Workflow Type: Galaxy
Protein target prediction of a bioactive ligand with Align-it and ePharmaLib
Associated Tutorial
This workflows is part of the tutorial Protein target prediction of a bioactive ligand with Align-it and ePharmaLib, available in the GTN
Thanks to...
Workflow Author(s): Aurélien F. A. Moumbock, Albert-Ludwigs-Universität Freiburg
Tutorial Author(s): Aurélien F. A. Moumbock, Simon Bray
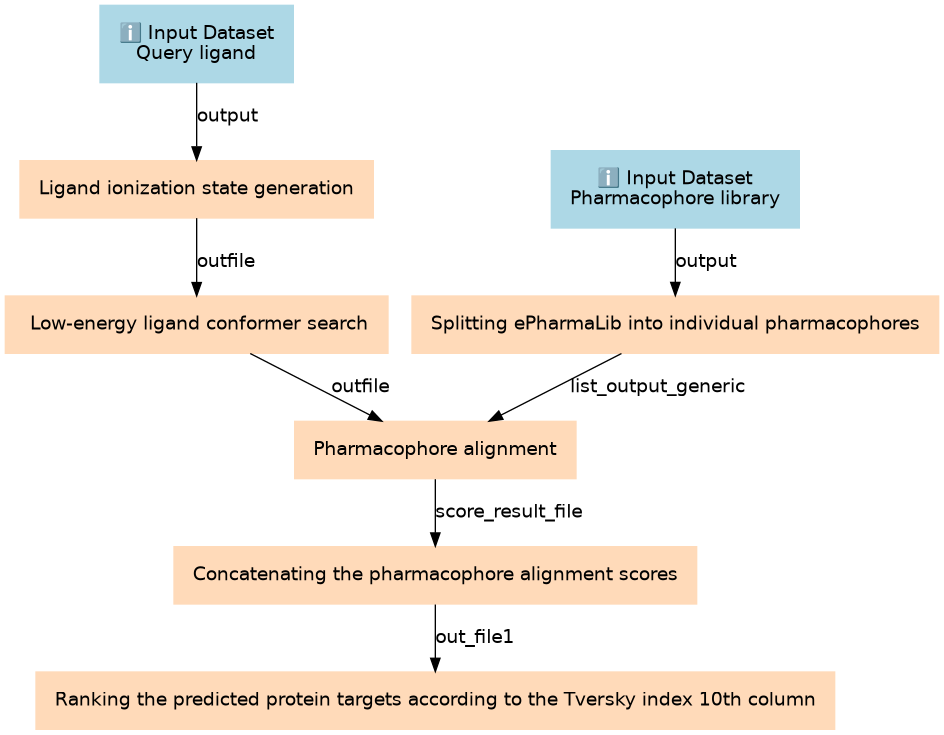
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Pharmacophore library | Pharmacophore library | n/a |
|
| Query ligand | Query ligand | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Ligand ionization state generation | toolshed.g2.bx.psu.edu/repos/bgruening/openbabel_addh/openbabel_addh/3.1.1+galaxy1 |
| 3 | Splitting ePharmaLib into individual pharmacophores | toolshed.g2.bx.psu.edu/repos/bgruening/split_file_to_collection/split_file_to_collection/0.5.0 |
| 4 | Low-energy ligand conformer search | toolshed.g2.bx.psu.edu/repos/bgruening/rdconf/rdconf/2020.03.4+galaxy0 |
| 5 | Pharmacophore alignment | toolshed.g2.bx.psu.edu/repos/bgruening/align_it/ctb_alignit/1.0.4+galaxy0 |
| 6 | Concatenating the pharmacophore alignment scores | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cat/0.1.1 |
| 7 | Ranking the predicted protein targets according to the Tversky index (10th column) | sort1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
| _anonymous_output_2 | _anonymous_output_2 | n/a |
|
| _anonymous_output_3 | _anonymous_output_3 | n/a |
|
| _anonymous_output_4 | _anonymous_output_4 | n/a |
|
| _anonymous_output_5 | _anonymous_output_5 | n/a |
|
| _anonymous_output_6 | _anonymous_output_6 | n/a |
|
| _anonymous_output_7 | _anonymous_output_7 | n/a |
|
| _anonymous_output_8 | _anonymous_output_8 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
License
Activity
Views: 237 Downloads: 35 Runs: 0
Created: 2nd Jun 2025 at 11:04
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master