Workflow Type: Galaxy
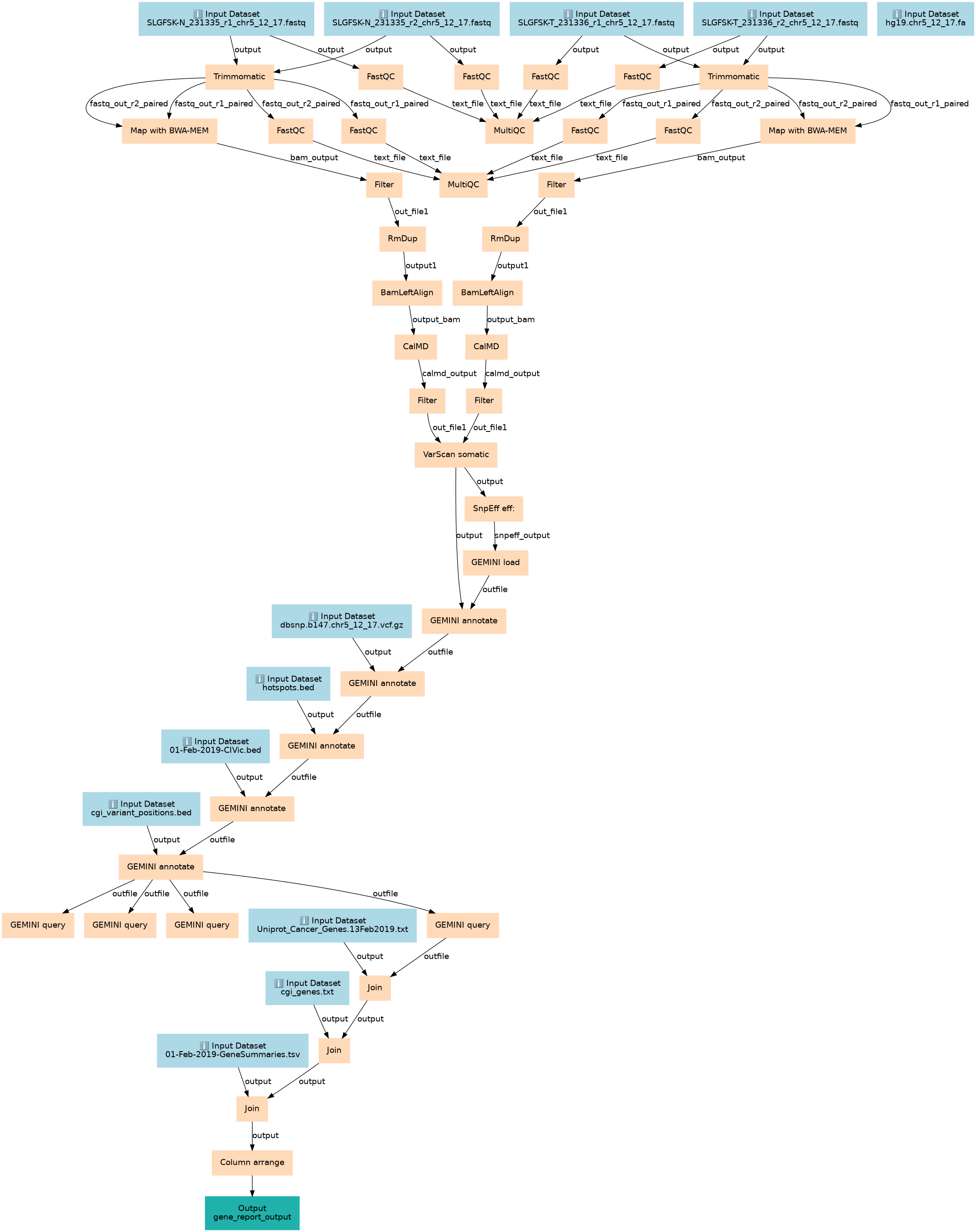
Identification of somatic and germline variants from tumor and normal sample pairs tutorial
Associated Tutorial
This workflows is part of the tutorial Identification of somatic and germline variants from tumor and normal sample pairs, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Tutorial Author(s): Wolfgang Maier
Tutorial Contributor(s): Verena Moosmann, Simon Gladman, Björn Grüning, Helena Rasche, Bérénice Batut, Wolfgang Maier, Saskia Hiltemann
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| 01-Feb-2019-CIVic.bed | 01-Feb-2019-CIVic.bed | n/a |
|
| 01-Feb-2019-GeneSummaries.tsv | 01-Feb-2019-GeneSummaries.tsv | n/a |
|
| SLGFSK-N_231335_r1_chr5_12_17.fastq | SLGFSK-N_231335_r1_chr5_12_17.fastq | n/a |
|
| SLGFSK-N_231335_r2_chr5_12_17.fastq | SLGFSK-N_231335_r2_chr5_12_17.fastq | n/a |
|
| SLGFSK-T_231336_r1_chr5_12_17.fastq | SLGFSK-T_231336_r1_chr5_12_17.fastq | n/a |
|
| SLGFSK-T_231336_r2_chr5_12_17.fastq | SLGFSK-T_231336_r2_chr5_12_17.fastq | n/a |
|
| Uniprot_Cancer_Genes.13Feb2019.txt | Uniprot_Cancer_Genes.13Feb2019.txt | n/a |
|
| cgi_genes.txt | cgi_genes.txt | n/a |
|
| cgi_variant_positions.bed | cgi_variant_positions.bed | n/a |
|
| dbsnp.b147.chr5_12_17.vcf.gz | dbsnp.b147.chr5_12_17.vcf.gz | n/a |
|
| hg19.chr5_12_17.fa | hg19.chr5_12_17.fa | n/a |
|
| hotspots.bed | hotspots.bed | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 12 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 13 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 14 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.36.5 |
| 15 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 16 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 17 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.36.5 |
| 18 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 19 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 20 | Map with BWA-MEM | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.17.1 |
| 21 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.8+galaxy0 |
| 22 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 23 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 24 | Map with BWA-MEM | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.17.1 |
| 25 | Filter | toolshed.g2.bx.psu.edu/repos/devteam/bamtools_filter/bamFilter/2.4.1 |
| 26 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.8+galaxy0 |
| 27 | Filter | toolshed.g2.bx.psu.edu/repos/devteam/bamtools_filter/bamFilter/2.4.1 |
| 28 | RmDup | toolshed.g2.bx.psu.edu/repos/devteam/samtools_rmdup/samtools_rmdup/2.0.1 |
| 29 | RmDup | toolshed.g2.bx.psu.edu/repos/devteam/samtools_rmdup/samtools_rmdup/2.0.1 |
| 30 | BamLeftAlign | toolshed.g2.bx.psu.edu/repos/devteam/freebayes/bamleftalign/1.3.1 |
| 31 | BamLeftAlign | toolshed.g2.bx.psu.edu/repos/devteam/freebayes/bamleftalign/1.3.1 |
| 32 | CalMD | toolshed.g2.bx.psu.edu/repos/devteam/samtools_calmd/samtools_calmd/2.0.2 |
| 33 | CalMD | toolshed.g2.bx.psu.edu/repos/devteam/samtools_calmd/samtools_calmd/2.0.2 |
| 34 | Filter | toolshed.g2.bx.psu.edu/repos/devteam/bamtools_filter/bamFilter/2.4.1 |
| 35 | Filter | toolshed.g2.bx.psu.edu/repos/devteam/bamtools_filter/bamFilter/2.4.1 |
| 36 | VarScan somatic | toolshed.g2.bx.psu.edu/repos/iuc/varscan_somatic/varscan_somatic/2.4.3.6 |
| 37 | SnpEff eff: | toolshed.g2.bx.psu.edu/repos/iuc/snpeff/snpEff/4.3+T.galaxy1 |
| 38 | GEMINI load | toolshed.g2.bx.psu.edu/repos/iuc/gemini_load/gemini_load/0.20.1+galaxy2 |
| 39 | GEMINI annotate | toolshed.g2.bx.psu.edu/repos/iuc/gemini_annotate/gemini_annotate/0.20.1+galaxy2 |
| 40 | GEMINI annotate | toolshed.g2.bx.psu.edu/repos/iuc/gemini_annotate/gemini_annotate/0.20.1+galaxy2 |
| 41 | GEMINI annotate | toolshed.g2.bx.psu.edu/repos/iuc/gemini_annotate/gemini_annotate/0.20.1+galaxy2 |
| 42 | GEMINI annotate | toolshed.g2.bx.psu.edu/repos/iuc/gemini_annotate/gemini_annotate/0.20.1+galaxy2 |
| 43 | GEMINI annotate | toolshed.g2.bx.psu.edu/repos/iuc/gemini_annotate/gemini_annotate/0.20.1+galaxy2 |
| 44 | GEMINI query | toolshed.g2.bx.psu.edu/repos/iuc/gemini_query/gemini_query/0.20.1+galaxy1 |
| 45 | GEMINI query | toolshed.g2.bx.psu.edu/repos/iuc/gemini_query/gemini_query/0.20.1+galaxy1 |
| 46 | GEMINI query | toolshed.g2.bx.psu.edu/repos/iuc/gemini_query/gemini_query/0.20.1+galaxy1 |
| 47 | GEMINI query | toolshed.g2.bx.psu.edu/repos/iuc/gemini_query/gemini_query/0.20.1+galaxy1 |
| 48 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/1.1.1 |
| 49 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/1.1.1 |
| 50 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/1.1.1 |
| 51 | Column arrange | toolshed.g2.bx.psu.edu/repos/bgruening/column_arrange_by_header/bg_column_arrange_by_header/0.2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| gene_report_output | gene_report_output | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 242 Downloads: 30 Runs: 0
Created: 2nd Jun 2025 at 11:03
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master