Workflow Type: Galaxy
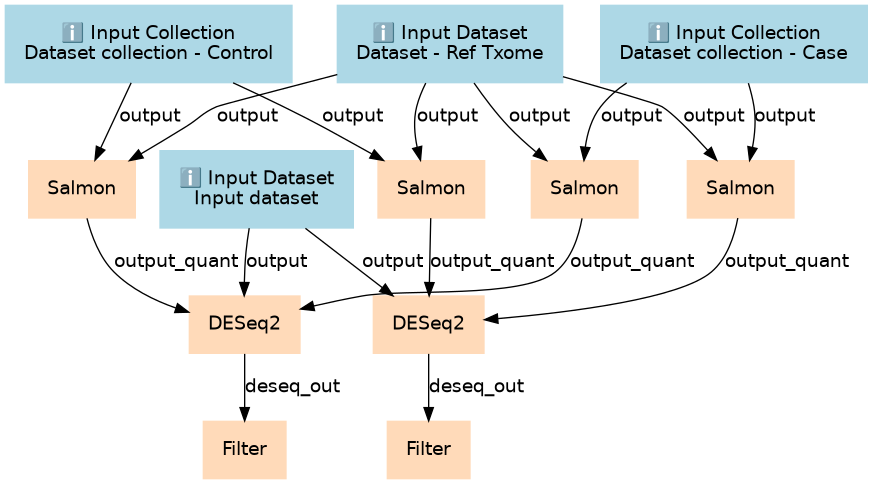
This workflow completes the second part of the sRNA-seq tutorial from transcript quantification through differential abundance testing.
Associated Tutorial
This workflows is part of the tutorial Differential abundance testing of small RNAs, available in the GTN
Thanks to...
Tutorial Author(s): Mallory Freeberg
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Salmon | Salmon quantification on control datasets calculating SF/sense hits toolshed.g2.bx.psu.edu/repos/bgruening/salmon/salmon/0.8.2 |
| 5 | Salmon | Salmon quantification on control datasets calculating SR/antisense hits toolshed.g2.bx.psu.edu/repos/bgruening/salmon/salmon/0.8.2 |
| 6 | Salmon | Salmon quantification on case datasets calculating SF/sense hits toolshed.g2.bx.psu.edu/repos/bgruening/salmon/salmon/0.8.2 |
| 7 | Salmon | Salmon quantification on case datasets calculating SR/antisense hits toolshed.g2.bx.psu.edu/repos/bgruening/salmon/salmon/0.8.2 |
| 8 | DESeq2 | Calculating differential abundance of SF/sense hits toolshed.g2.bx.psu.edu/repos/iuc/deseq2/deseq2/2.11.39 |
| 9 | DESeq2 | Calculating differential abundance of SR/antisense hits toolshed.g2.bx.psu.edu/repos/iuc/deseq2/deseq2/2.11.39 |
| 10 | Filter | Filter1 |
| 11 | Filter | Filter1 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 227 Downloads: 30 Runs: 0
Created: 2nd Jun 2025 at 11:03
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master