Workflow Type: Galaxy
Secondary metabolite biosynthetic gene cluster (SMBGC) Annotation using Neural Networks Trained on Interpro Signatures
Associated Tutorial
This workflows is part of the tutorial Marine Omics identifying biosynthetic gene clusters, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Marie Jossé
Tutorial Author(s): Marie Josse
Tutorial Contributor(s): Björn Grüning, Saskia Hiltemann
Grants(s): Fair-Ease, EuroScienceGateway
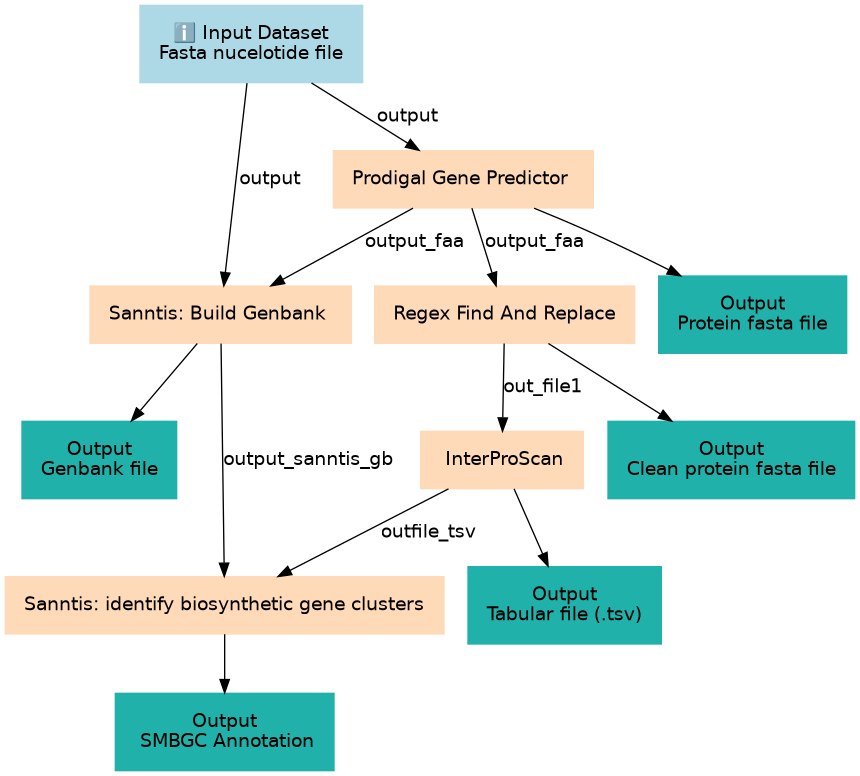
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Fasta nucelotide file | Fasta nucelotide file | BGC0001472.fna |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | Prodigal Gene Predictor | Create the protein fasta file toolshed.g2.bx.psu.edu/repos/iuc/prodigal/prodigal/2.6.3+galaxy0 |
| 2 | Sanntis: Build Genbank | Use of Sanntis toolshed.g2.bx.psu.edu/repos/ecology/sanntis_marine/sanntis_marine/0.9.3.5+galaxy1 |
| 3 | Regex Find And Replace | Remove useless * in the protein fasta file toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regex1/1.0.3 |
| 4 | InterProScan | Create TSV file for Sanntis toolshed.g2.bx.psu.edu/repos/bgruening/interproscan/interproscan/5.59-91.0+galaxy3 |
| 5 | Sanntis: identify biosynthetic gene clusters | toolshed.g2.bx.psu.edu/repos/ecology/sanntis_marine/sanntis_marine/0.9.3.5+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Protein fasta file | Protein fasta file | n/a |
|
| Genbank file | Genbank file | n/a |
|
| Clean protein fasta file | Clean protein fasta file | n/a |
|
| Tabular file (.tsv) | Tabular file (.tsv) | n/a |
|
| SMBGC Annotation | SMBGC Annotation | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 255 Downloads: 38 Runs: 0
Created: 2nd Jun 2025 at 11:03
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master