Workflow Type: Galaxy
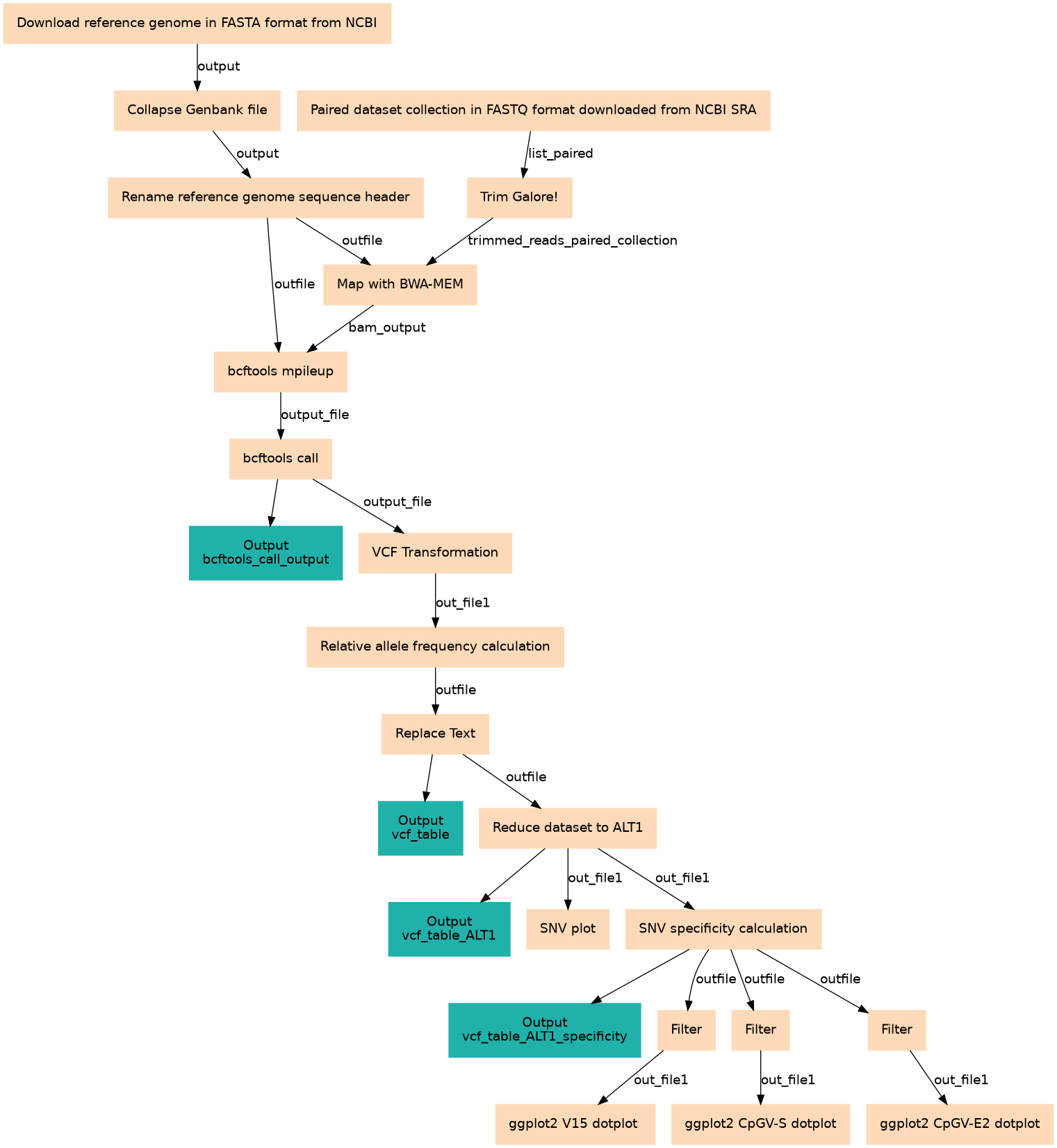
This standard workflow determines SNV positions from baculovirus isolates using bcftools.
Associated Tutorial
This workflows is part of the tutorial Deciphering Virus Populations - Single Nucleotide Variants (SNVs) and Specificities in Baculovirus Isolates, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Jörg T. Wennmann
Tutorial Author(s): Jörg Wennmann
Steps
| ID | Name | Description |
|---|---|---|
| 0 | Download reference genome in FASTA format from NCBI | Exactly one reference genome. toolshed.g2.bx.psu.edu/repos/iuc/ncbi_acc_download/ncbi_acc_download/0.2.8+galaxy0 |
| 1 | Paired dataset collection in FASTQ format (downloaded from NCBI SRA) | Provide a dataset list of paired (forward and reverse) reads in fastq format. toolshed.g2.bx.psu.edu/repos/iuc/sra_tools/fastq_dump/3.1.1+galaxy1 |
| 2 | Collapse Genbank file | NCBI Download accepts multiple accession numbers and creates a list as output. Therefore, the Fasta files can contain multiple records. For this analysis just one reference genome is required and the list needs to be converted into one file. toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 3 | Trim Galore! | toolshed.g2.bx.psu.edu/repos/bgruening/trim_galore/trim_galore/0.6.7+galaxy0 |
| 4 | Rename reference genome sequence header | Provide a descriptive and unique name for the reference sequence. toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_line/9.3+galaxy1 |
| 5 | Map with BWA-MEM | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.18 |
| 6 | bcftools mpileup | toolshed.g2.bx.psu.edu/repos/iuc/bcftools_mpileup/bcftools_mpileup/1.15.1+galaxy4 |
| 7 | bcftools call | toolshed.g2.bx.psu.edu/repos/iuc/bcftools_call/bcftools_call/1.15.1+galaxy5 |
| 8 | VCF Transformation | The VCF is converted to an easier to read table format. toolshed.g2.bx.psu.edu/repos/devteam/vcf2tsv/vcf2tsv/1.0.0_rc1+galaxy0 |
| 9 | Relative allele frequency calculation | The DPR value is further decomposed for each position and isolate and attached to the table. Four additional columns are added to the table: ALLELE, DPR.ALLELE, REL.ALT and REL.ALT.0.05. toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_awk_tool/9.3+galaxy1 |
| 10 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1 |
| 11 | Reduce dataset to ALT1 | In this step the dataset is reduced to the first alternative nucleotide (ALT1) only. Filter1 |
| 12 | SNV plot | A general overview to access the isolates overall heterogeneity/homogeneity. toolshed.g2.bx.psu.edu/repos/iuc/ggplot2_point/ggplot2_point/3.4.0+galaxy1 |
| 13 | SNV specificity calculation | The DPR value is further decomposed for each position and sample and attached to the table. Four additional columns are added to the table: ALLELE, DPR.ALLELE, REL.ALT and REL.ALT.0.05. toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_awk_tool/9.3+galaxy1 |
| 14 | Filter | Filter1 |
| 15 | Filter | Filter1 |
| 16 | Filter | Filter1 |
| 17 | ggplot2 V15 dotplot | Creates a ggplot2 SNV plot for each SNV specificity. toolshed.g2.bx.psu.edu/repos/iuc/ggplot2_point/ggplot2_point/3.4.0+galaxy1 |
| 18 | ggplot2 CpGV-S dotplot | Creates a ggplot2 SNV plot for each SNV specificity. toolshed.g2.bx.psu.edu/repos/iuc/ggplot2_point/ggplot2_point/3.4.0+galaxy1 |
| 19 | ggplot2 CpGV-E2 dotplot | Creates a ggplot2 SNV plot for each SNV specificity. toolshed.g2.bx.psu.edu/repos/iuc/ggplot2_point/ggplot2_point/3.4.0+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| bcftools_call_output | bcftools_call_output | n/a |
|
| vcf_table | vcf_table | n/a |
|
| vcf_table_ALT1 | vcf_table_ALT1 | n/a |
|
| vcf_table_ALT1_specificity | vcf_table_ALT1_specificity | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 226 Downloads: 28 Runs: 0
Created: 2nd Jun 2025 at 11:01
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master