Workflow Type: Galaxy
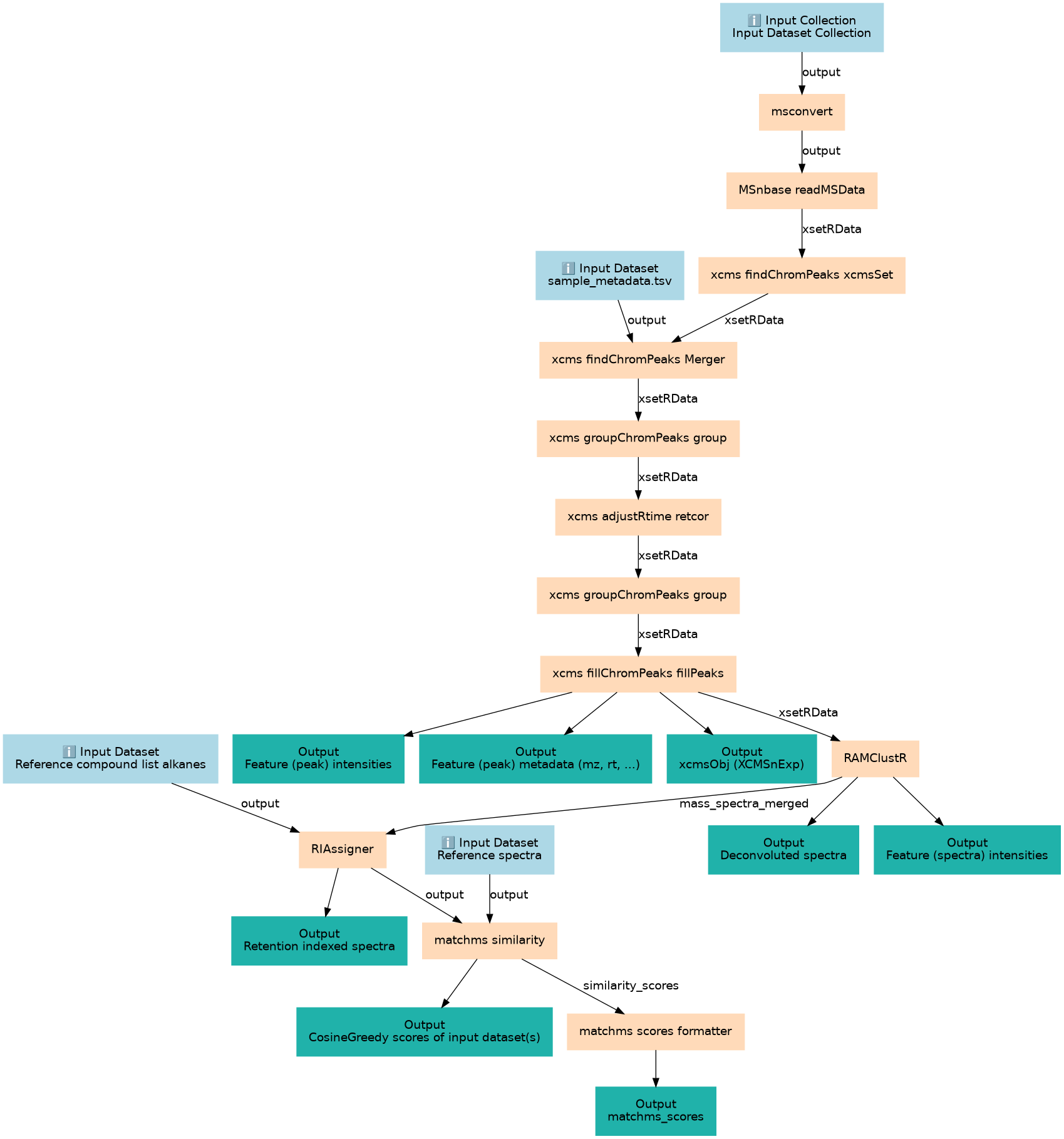
XCMS and RAMClustR based workflow for data processing and annotation using library matching via matchms.
Associated Tutorial
This workflows is part of the tutorial Mass spectrometry: GC-MS data processing (with XCMS, RAMClustR, RIAssigner, and matchms), available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): RECETOX
Tutorial Author(s): Matej Troják, Helge Hecht, Maxim Skoryk
Tutorial Contributor(s): Helena Rasche, Matej Troják, Mélanie Petera, Saskia Hiltemann, Björn Grüning
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Input Dataset Collection | #main/Input Dataset Collection | Mass spectrometry data to be processed in the workflow. Please ensure the format is supported by msconvert and only use the peak picking option if the input data was acquired in profile mode. |
|
| Reference compound list (alkanes) | #main/Reference compound list (alkanes) | List of retention indexed reference compounds. Either a table with columns `rt` and `ri` or an MSP with retention time and index metadata. |
|
| Reference spectra | #main/Reference spectra | Spectral library to use for annotation in MSP or MGF format. |
|
| sample_metadata.tsv | #main/sample_metadata.tsv | Sample metadata sheet, containing sample name, type (QC, blank, sample, etc.), batch number and injection order. |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | msconvert | toolshed.g2.bx.psu.edu/repos/galaxyp/msconvert/msconvert/3.0.20287.2 |
| 5 | MSnbase readMSData | toolshed.g2.bx.psu.edu/repos/lecorguille/msnbase_readmsdata/msnbase_readmsdata/2.16.1+galaxy0 |
| 6 | xcms findChromPeaks (xcmsSet) | toolshed.g2.bx.psu.edu/repos/lecorguille/xcms_xcmsset/abims_xcms_xcmsSet/3.12.0+galaxy0 |
| 7 | xcms findChromPeaks Merger | toolshed.g2.bx.psu.edu/repos/lecorguille/xcms_merge/xcms_merge/3.12.0+galaxy0 |
| 8 | xcms groupChromPeaks (group) | toolshed.g2.bx.psu.edu/repos/lecorguille/xcms_group/abims_xcms_group/3.12.0+galaxy0 |
| 9 | xcms adjustRtime (retcor) | toolshed.g2.bx.psu.edu/repos/lecorguille/xcms_retcor/abims_xcms_retcor/3.12.0+galaxy0 |
| 10 | xcms groupChromPeaks (group) | toolshed.g2.bx.psu.edu/repos/lecorguille/xcms_group/abims_xcms_group/3.12.0+galaxy0 |
| 11 | xcms fillChromPeaks (fillPeaks) | toolshed.g2.bx.psu.edu/repos/lecorguille/xcms_fillpeaks/abims_xcms_fillPeaks/3.12.0+galaxy0 |
| 12 | RAMClustR | toolshed.g2.bx.psu.edu/repos/recetox/ramclustr/ramclustr/1.2.4+galaxy2 |
| 13 | RIAssigner | toolshed.g2.bx.psu.edu/repos/recetox/riassigner/riassigner/0.3.4+galaxy1 |
| 14 | matchms similarity | toolshed.g2.bx.psu.edu/repos/recetox/matchms_similarity/matchms_similarity/0.20.0+galaxy0 |
| 15 | matchms scores formatter | toolshed.g2.bx.psu.edu/repos/recetox/matchms_formatter/matchms_formatter/0.20.0+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| CosineGreedy scores of input dataset(s) | #main/CosineGreedy scores of input dataset(s) | n/a |
|
| Deconvoluted spectra | #main/Deconvoluted spectra | n/a |
|
| Feature (peak) intensities | #main/Feature (peak) intensities | n/a |
|
| Feature (peak) metadata (mz, rt, ...) | #main/Feature (peak) metadata (mz, rt, ...) | n/a |
|
| Feature (spectra) intensities | #main/Feature (spectra) intensities | n/a |
|
| Retention indexed spectra | #main/Retention indexed spectra | n/a |
|
| matchms_scores | #main/matchms_scores | n/a |
|
| xcmsObj (XCMSnExp) | #main/xcmsObj (XCMSnExp) | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 227 Downloads: 31 Runs: 0
Created: 2nd Jun 2025 at 10:59
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master