Workflow Type: Galaxy
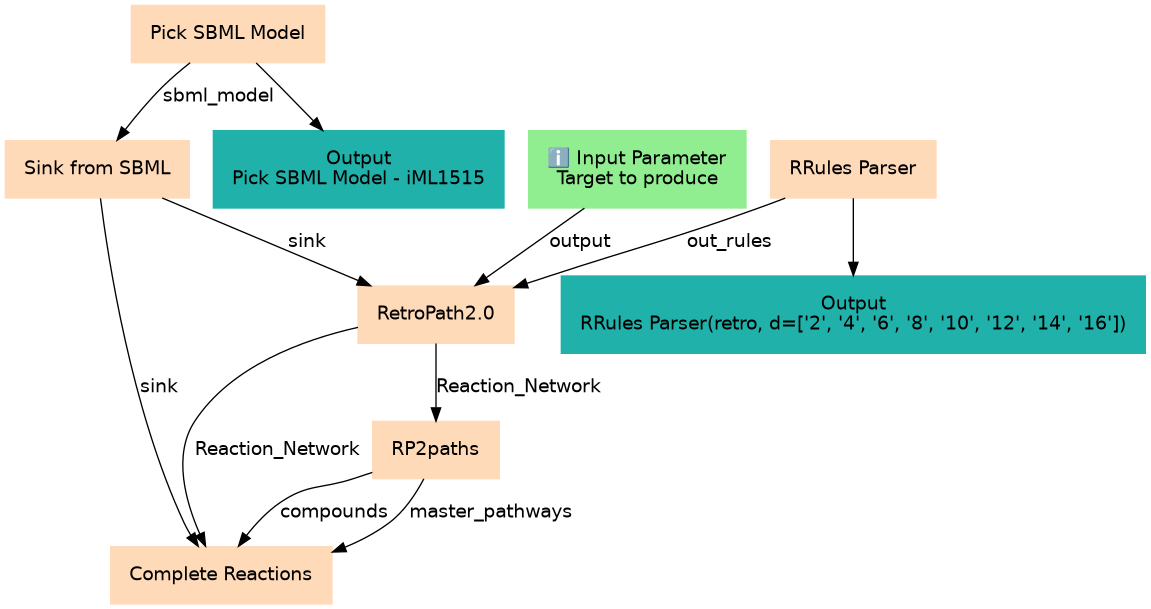
Generating theoretical possible pathways for the production of Lycopene in E.Coli using Retrosynthesis tools
Associated Tutorial
This workflows is part of the tutorial Generating theoretical possible pathways for the production of Lycopene in E.Coli using Retrosynthesis tools, available in the GTN
Thanks to...
Tutorial Author(s): Guillaume Gricourt, Thomas Duigou, Kenza Bazi-Kabbaj, Joan Hérisson, Ioana Popescu, Jean-Loup Faulon
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Target to produce | #main/Target to produce | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | Pick SBML Model | toolshed.g2.bx.psu.edu/repos/tduigou/get_sbml_model/get_sbml_model/0.0.1 |
| 2 | RRules Parser | toolshed.g2.bx.psu.edu/repos/tduigou/rrparser/rrparser/2.4.6 |
| 3 | Sink from SBML | toolshed.g2.bx.psu.edu/repos/tduigou/rpextractsink/rpextractsink/5.12.1 |
| 4 | RetroPath2.0 | toolshed.g2.bx.psu.edu/repos/tduigou/retropath2/retropath2/2.3.0 |
| 5 | RP2paths | toolshed.g2.bx.psu.edu/repos/tduigou/rp2paths/rp2paths/1.5.0 |
| 6 | Complete Reactions | toolshed.g2.bx.psu.edu/repos/tduigou/rpcompletion/rpcompletion/5.12.2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Pick SBML Model - iML1515 | #main/Pick SBML Model - iML1515 | n/a |
|
| RRules Parser(retro, d=['2', '4', '6', '8', '10', '12', '14', '16']) | #main/RRules Parser(retro, d=['2', '4', '6', '8', '10', '12', '14', '16']) | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 226 Downloads: 32 Runs: 0
Created: 2nd Jun 2025 at 10:59
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master