Workflow Type: Galaxy
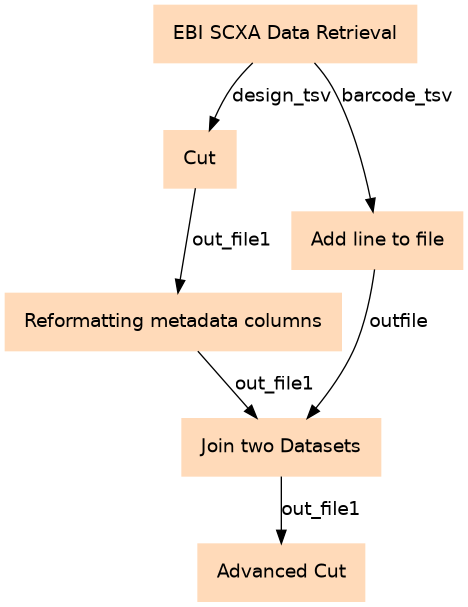
This workflow generates from only an EBI SCXA reference the metadata for creating an ESet object
Associated Tutorial
This workflows is part of the tutorial Matrix Exchange Format to ESet | Creating a single-cell RNA-seq reference dataset for deconvolution, available in the GTN
Thanks to...
Workflow Author(s): Wendi Bacon, Mehmet Tekman
Tutorial Author(s): Wendi Bacon, Mehmet Tekman
Tutorial Contributor(s): Marisa Loach, Mehmet Tekman, Saskia Hiltemann, Wendi Bacon, Helena Rasche, Pablo Moreno, Julia Jakiela, Pavankumar Videm
Steps
| ID | Name | Description |
|---|---|---|
| 0 | EBI SCXA Data Retrieval | toolshed.g2.bx.psu.edu/repos/ebi-gxa/retrieve_scxa/retrieve_scxa/v0.0.1+galaxy1 |
| 1 | Cut | Cut1 |
| 2 | Add line to file | toolshed.g2.bx.psu.edu/repos/bgruening/add_line_to_file/add_line_to_file/0.1.0 |
| 3 | Reformatting metadata columns | Adjust this step as necessary toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regex1/1.0.2 |
| 4 | Join two Datasets | Experimental designs often include extra stuff (likely the barcodes not meeting the EBI pre-processing criteria), so this way we streamline down to only those barcodes in the barcodes file. join1 |
| 5 | Advanced Cut | Gets rid of the duplicated columns toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/1.1.0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| _anonymous_output_8 | #main/_anonymous_output_8 | n/a |
|
| _anonymous_output_9 | #main/_anonymous_output_9 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 236 Downloads: 31 Runs: 0
Created: 2nd Jun 2025 at 10:58
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master