Workflow Type: Galaxy
Associated Tutorial
This workflows is part of the tutorial Pre-processing of 10X Single-Cell RNA Datasets, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Wendi Bacon, Pavankumar Videm, Mehmet Tekman, Hans-Rudolf Hotz, Daniel Blankenberg
Tutorial Author(s): Mehmet Tekman, Hans-Rudolf Hotz, Daniel Blankenberg, Pavankumar Videm
Tutorial Contributor(s): Wendi Bacon, Björn Grüning, Donny Vrins, Saskia Hiltemann, Helena Rasche, Pavankumar Videm, Mehmet Tekman, Bérénice Batut, Hans-Rudolf Hotz, Cristóbal Gallardo
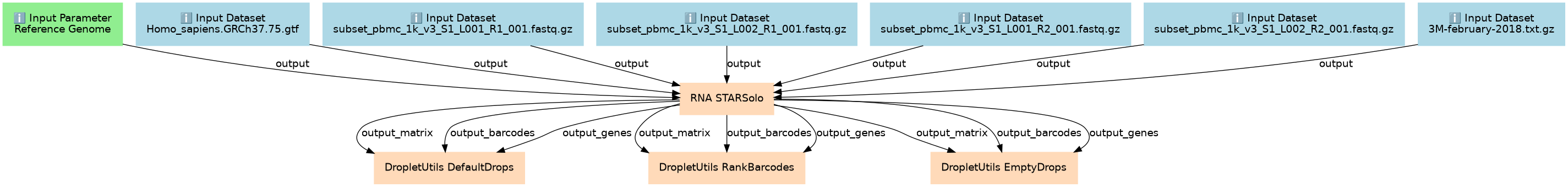
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| 3M-february-2018.txt.gz | 3M-february-2018.txt.gz | n/a |
|
| Homo_sapiens.GRCh37.75.gtf | Homo_sapiens.GRCh37.75.gtf | n/a |
|
| Reference Genome | Reference Genome | n/a |
|
| subset_pbmc_1k_v3_S1_L001_R1_001.fastq.gz | subset_pbmc_1k_v3_S1_L001_R1_001.fastq.gz | n/a |
|
| subset_pbmc_1k_v3_S1_L001_R2_001.fastq.gz | subset_pbmc_1k_v3_S1_L001_R2_001.fastq.gz | n/a |
|
| subset_pbmc_1k_v3_S1_L002_R1_001.fastq.gz | subset_pbmc_1k_v3_S1_L002_R1_001.fastq.gz | n/a |
|
| subset_pbmc_1k_v3_S1_L002_R2_001.fastq.gz | subset_pbmc_1k_v3_S1_L002_R2_001.fastq.gz | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 7 | RNA STARSolo | toolshed.g2.bx.psu.edu/repos/iuc/rna_starsolo/rna_starsolo/2.7.11a+galaxy1 |
| 8 | DropletUtils DefaultDrops | toolshed.g2.bx.psu.edu/repos/iuc/dropletutils/dropletutils/1.10.0+galaxy2 |
| 9 | DropletUtils RankBarcodes | toolshed.g2.bx.psu.edu/repos/iuc/dropletutils/dropletutils/1.10.0+galaxy2 |
| 10 | DropletUtils EmptyDrops | toolshed.g2.bx.psu.edu/repos/iuc/dropletutils/dropletutils/1.10.0+galaxy2 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 227 Downloads: 33 Runs: 0
Created: 2nd Jun 2025 at 10:58
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master