Workflow Type: Galaxy
Creates input file for Filter, Plot, Explore tutorial
Associated Tutorial
This workflows is part of the tutorial Importing files from public atlases, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Julia Jakiela
Tutorial Author(s): Julia Jakiela, Wendi Bacon
Tutorial Contributor(s): Björn Grüning, Julia Jakiela, Wendi Bacon, Helena Rasche, Pablo Moreno, Pavankumar Videm, Mehmet Tekman, Saskia Hiltemann
Grants(s): DASH UK
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| SC-Atlas experiment accession | SC-Atlas experiment accession | EBI Single Cell Atlas accession for the experiment that you want to retrieve. |
|
Steps
| ID | Name | Description |
|---|---|---|
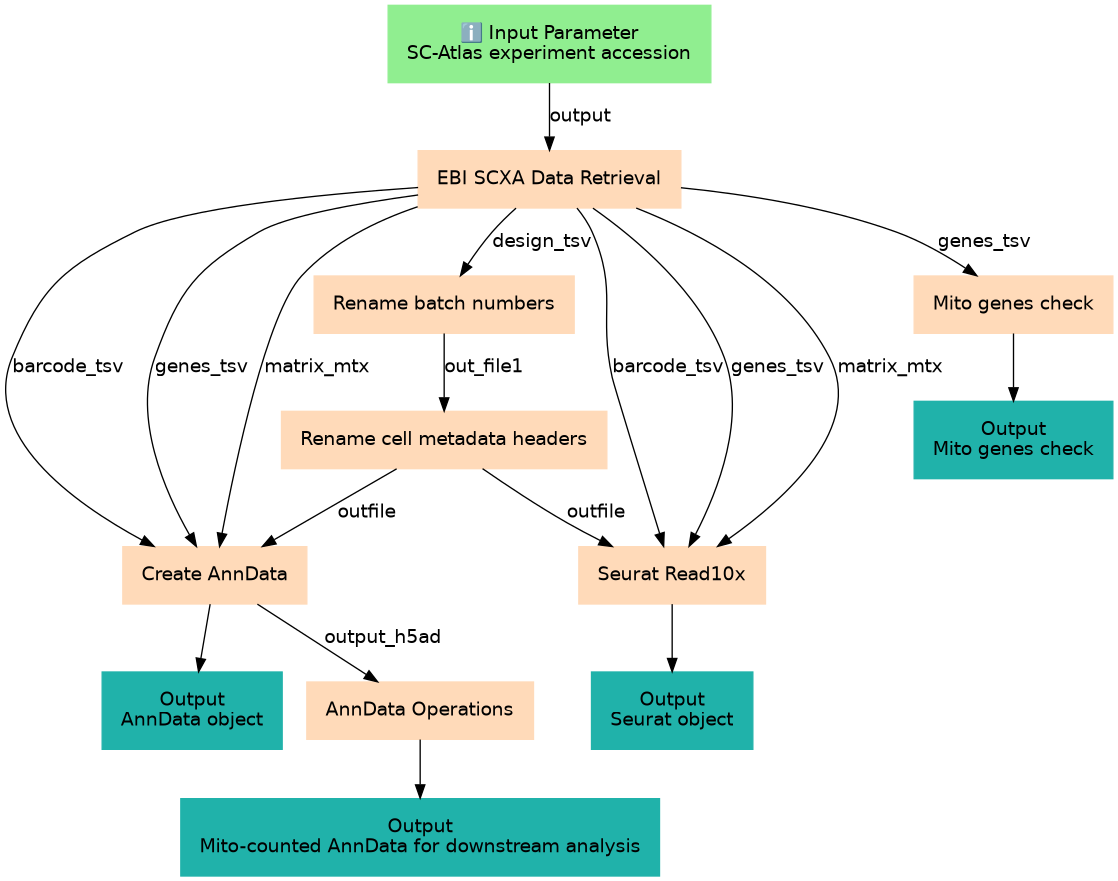
| 1 | EBI SCXA Data Retrieval | toolshed.g2.bx.psu.edu/repos/ebi-gxa/retrieve_scxa/retrieve_scxa/v0.0.2+galaxy2 |
| 2 | Rename batch numbers | Changes the batch label from "1" to "N01" etc toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.3 |
| 3 | Mito genes check | Shows how mitochondrial genes are annotated. toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/9.3+galaxy1 |
| 4 | Rename cell metadata headers | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_find_and_replace/9.3+galaxy1 |
| 5 | Create AnnData | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_read_10x/scanpy_read_10x/1.9.3+galaxy0 |
| 6 | Seurat Read10x | toolshed.g2.bx.psu.edu/repos/ebi-gxa/seurat_read10x/seurat_read10x/4.0.4+galaxy0 |
| 7 | AnnData Operations | toolshed.g2.bx.psu.edu/repos/ebi-gxa/anndata_ops/anndata_ops/1.9.3+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Mito genes check | Mito genes check | n/a |
|
| AnnData object | AnnData object | n/a |
|
| Seurat object | Seurat object | n/a |
|
| Mito-counted AnnData for downstream analysis | Mito-counted AnnData for downstream analysis | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tool
Activity
Views: 235 Downloads: 34 Runs: 0
Created: 2nd Jun 2025 at 10:57
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master