Workflow Type: Galaxy
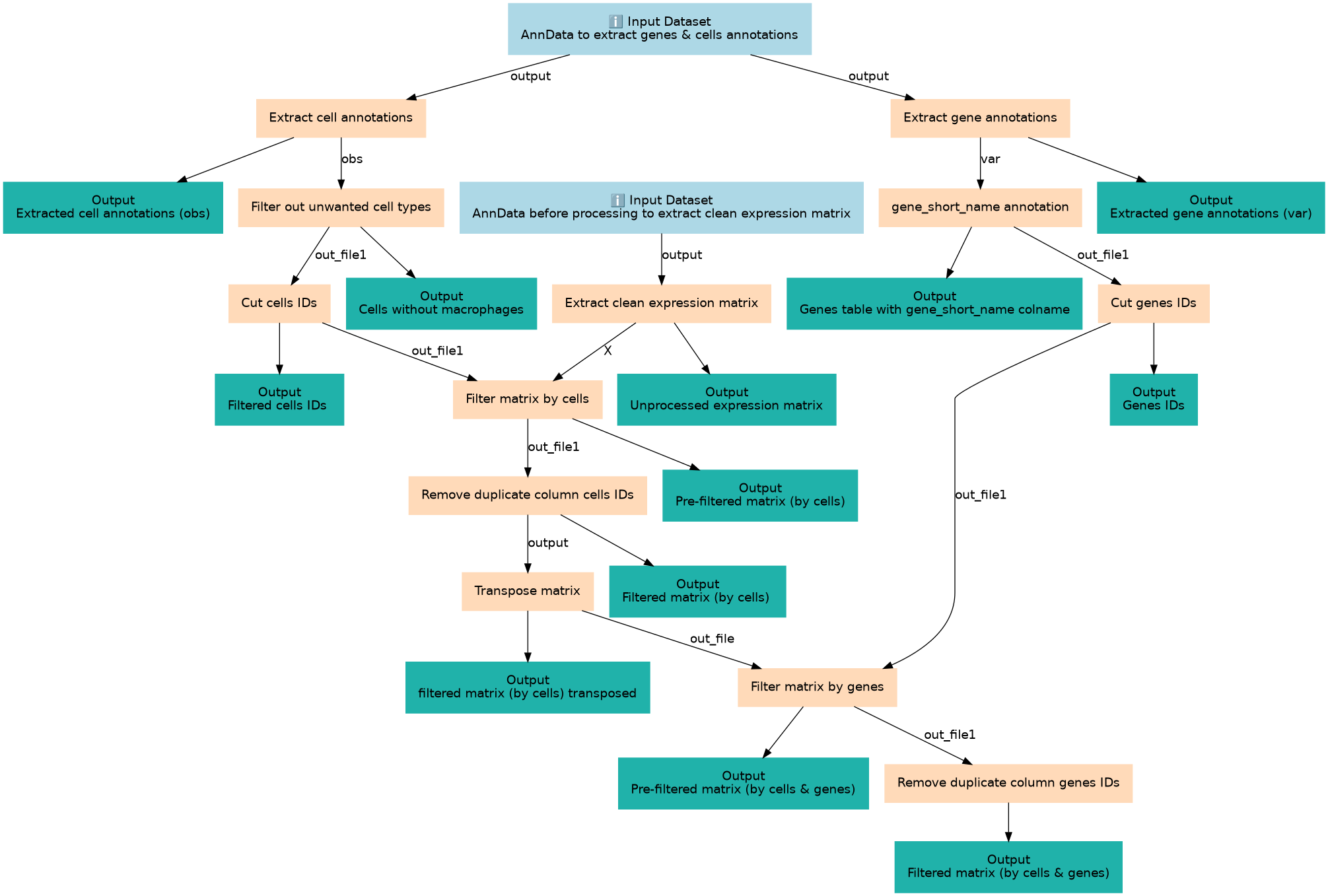
Preparing and filtering gene and cell annotations files and expression matrix to be passed as input for Monocle
Associated Tutorial
This workflows is part of the tutorial Inferring single cell trajectories with Monocle3, available in the GTN
Thanks to...
Tutorial Author(s): Julia Jakiela
Tutorial Contributor(s): Helena Rasche, Wendi Bacon, Pavankumar Videm, Julia Jakiela, Saskia Hiltemann, Mehmet Tekman, Matthias Bernt, Pablo Moreno, Björn Grüning
Grants(s): EPSRC Training Grant DTP 2020-2021 Open University
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| AnnData before processing to extract clean expression matrix | #main/AnnData before processing to extract clean expression matrix | n/a |
|
| AnnData to extract genes & cells annotations | #main/AnnData to extract genes & cells annotations | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Extract cell annotations | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 3 | Extract gene annotations | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 4 | Extract clean expression matrix | Unprocessed means here before normalisation or dimensionality reduction. For this step, must have cell IDs as rownames. toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 5 | Filter out unwanted cell types | Double-check the cell_type column number Filter1 |
| 6 | gene_short_name annotation | Check the number of the column with genes names (using column) and its header (Find Regex) toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.2 |
| 7 | Cut cells IDs | Cut1 |
| 8 | Cut genes IDs | Cut1 |
| 9 | Filter matrix (by cells) | join1 |
| 10 | Remove duplicate column (cells IDs) | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/1.1.0 |
| 11 | Transpose matrix | toolshed.g2.bx.psu.edu/repos/iuc/datamash_transpose/datamash_transpose/1.1.0+galaxy2 |
| 12 | Filter matrix (by genes) | join1 |
| 13 | Remove duplicate column (genes IDs) | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/1.1.0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Cells without macrophages | #main/Cells without macrophages | n/a |
|
| Extracted cell annotations (obs) | #main/Extracted cell annotations (obs) | n/a |
|
| Extracted gene annotations (var) | #main/Extracted gene annotations (var) | n/a |
|
| Filtered cells IDs | #main/Filtered cells IDs | n/a |
|
| Filtered matrix (by cells & genes) | #main/Filtered matrix (by cells & genes) | n/a |
|
| Filtered matrix (by cells) | #main/Filtered matrix (by cells) | n/a |
|
| Genes IDs | #main/Genes IDs | n/a |
|
| Genes table with gene_short_name colname | #main/Genes table with gene_short_name colname | n/a |
|
| Pre-filtered matrix (by cells & genes) | #main/Pre-filtered matrix (by cells & genes) | n/a |
|
| Pre-filtered matrix (by cells) | #main/Pre-filtered matrix (by cells) | n/a |
|
| Unprocessed expression matrix | #main/Unprocessed expression matrix | n/a |
|
| filtered matrix (by cells) transposed | #main/filtered matrix (by cells) transposed | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tool
Activity
Views: 232 Downloads: 33 Runs: 0
Created: 2nd Jun 2025 at 10:56
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master