Workflow Type: Galaxy
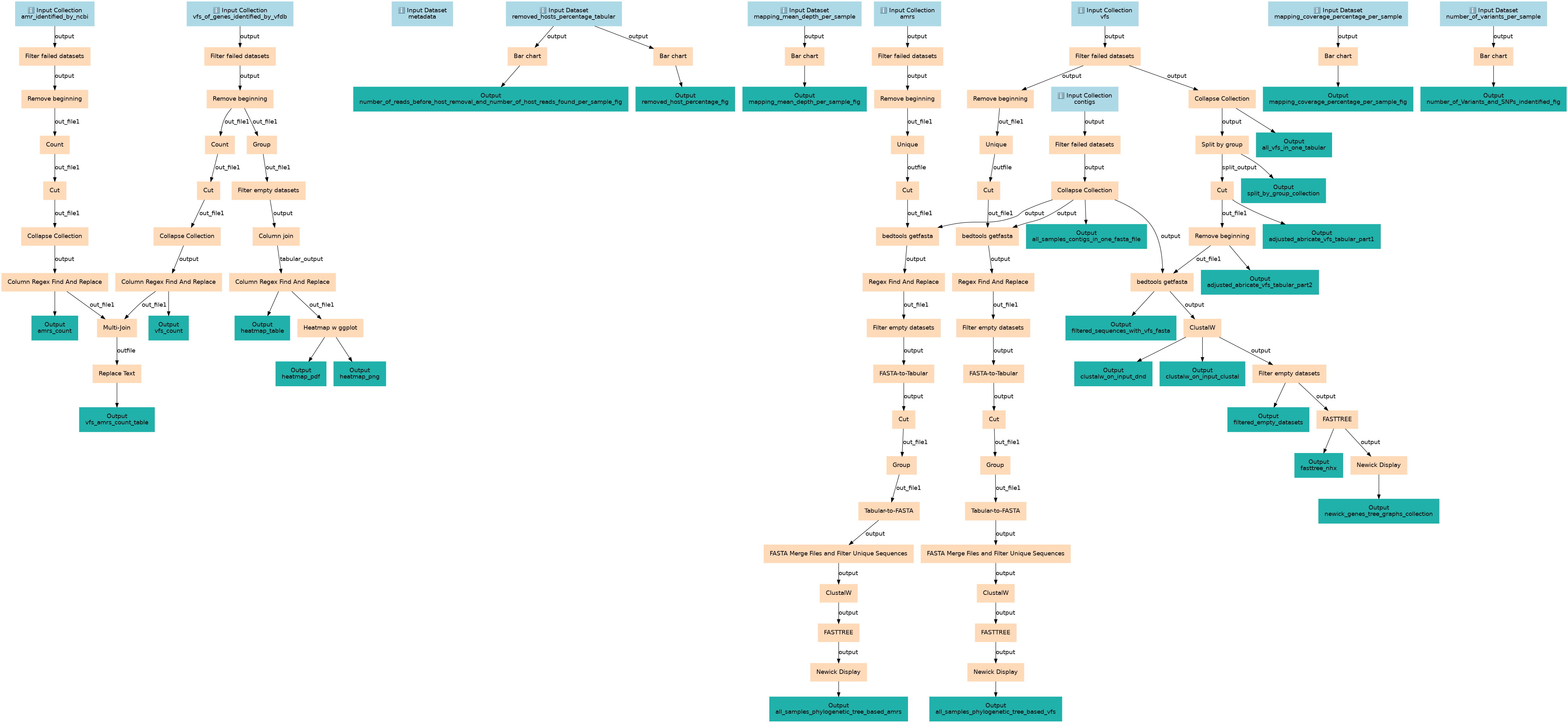
Pathogens of all samples report generation and visualization
Associated Tutorial
This workflows is part of the tutorial Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition, available in the GTN
Features
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Engy Nasr, Bérénice Batut, Paul Zierep
Tutorial Author(s): Bérénice Batut, Engy Nasr, Paul Zierep
Tutorial Contributor(s): Hans-Rudolf Hotz, Wolfgang Maier, Saskia Hiltemann, Deepti Varshney, Paul Zierep, Bérénice Batut, Björn Grüning, Cristóbal Gallardo, Engy Nasr, Helena Rasche
Grants(s): Gallantries: Bridging Training Communities in Life Science, Environment and Health, EOSC-Life
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| amr_identified_by_ncbi | amr_identified_by_ncbi | output_of_abricate_with_amrfinderncbi |
|
| amrs | amrs | amrs |
|
| contigs | contigs | Contigs |
|
| mapping_coverage_percentage_per_sample | mapping_coverage_percentage_per_sample | mapping_coverage_percentage_per_sample |
|
| mapping_mean_depth_per_sample | mapping_mean_depth_per_sample | mapping_mean_depth_per_sample |
|
| metadata | metadata | samples_metadata |
|
| number_of_variants_per_sample | number_of_variants_per_sample | number_of_variants_per_sample |
|
| removed_hosts_percentage_tabular | removed_hosts_percentage_tabular | removed_hosts_percentage_tabular |
|
| vfs | vfs | VFs |
|
| vfs_of_genes_identified_by_vfdb | vfs_of_genes_identified_by_vfdb | output_of_abricate_with_vfdb |
|
Steps
| ID | Name | Description |
|---|---|---|
| 10 | Filter failed datasets | __FILTER_FAILED_DATASETS__ |
| 11 | Filter failed datasets | __FILTER_FAILED_DATASETS__ |
| 12 | Bar chart | barchart_gnuplot |
| 13 | Bar chart | barchart_gnuplot |
| 14 | Bar chart | barchart_gnuplot |
| 15 | Filter failed datasets | __FILTER_FAILED_DATASETS__ |
| 16 | Bar chart | barchart_gnuplot |
| 17 | Bar chart | barchart_gnuplot |
| 18 | Filter failed datasets | __FILTER_FAILED_DATASETS__ |
| 19 | Filter failed datasets | __FILTER_FAILED_DATASETS__ |
| 20 | Remove beginning | Remove beginning1 |
| 21 | Remove beginning | Remove beginning1 |
| 22 | Remove beginning | Remove beginning1 |
| 23 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 24 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 25 | Remove beginning | Remove beginning1 |
| 26 | Count | Count1 |
| 27 | Count | Count1 |
| 28 | Group | Grouping1 |
| 29 | Unique | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sorted_uniq/9.3+galaxy1 |
| 30 | Split by group | toolshed.g2.bx.psu.edu/repos/bgruening/split_file_on_column/tp_split_on_column/0.6 |
| 31 | Unique | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sorted_uniq/9.3+galaxy1 |
| 32 | Cut | Cut1 |
| 33 | Cut | Cut1 |
| 34 | Filter empty datasets | __FILTER_EMPTY_DATASETS__ |
| 35 | Cut | Cut1 |
| 36 | Cut | Cut1 |
| 37 | Cut | Cut1 |
| 38 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 39 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 40 | Column join | toolshed.g2.bx.psu.edu/repos/iuc/collection_column_join/collection_column_join/0.0.3 |
| 41 | bedtools getfasta | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_getfastabed/2.30.0+galaxy1 |
| 42 | Remove beginning | Remove beginning1 |
| 43 | bedtools getfasta | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_getfastabed/2.30.0+galaxy1 |
| 44 | Column Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.3 |
| 45 | Column Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.3 |
| 46 | Column Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.3 |
| 47 | Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regex1/1.0.3 |
| 48 | bedtools getfasta | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_getfastabed/2.30.0+galaxy1 |
| 49 | Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regex1/1.0.3 |
| 50 | Multi-Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_multijoin_tool/9.3+galaxy1 |
| 51 | Heatmap w ggplot | toolshed.g2.bx.psu.edu/repos/iuc/ggplot2_heatmap/ggplot2_heatmap/3.4.0+galaxy0 |
| 52 | Filter empty datasets | __FILTER_EMPTY_DATASETS__ |
| 53 | ClustalW | toolshed.g2.bx.psu.edu/repos/devteam/clustalw/clustalw/2.1+galaxy1 |
| 54 | Filter empty datasets | __FILTER_EMPTY_DATASETS__ |
| 55 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1 |
| 56 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.1 |
| 57 | Filter empty datasets | __FILTER_EMPTY_DATASETS__ |
| 58 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.1 |
| 59 | Cut | Cut1 |
| 60 | FASTTREE | toolshed.g2.bx.psu.edu/repos/iuc/fasttree/fasttree/2.1.10+galaxy1 |
| 61 | Cut | Cut1 |
| 62 | Group | Grouping1 |
| 63 | Newick Display | toolshed.g2.bx.psu.edu/repos/iuc/newick_utils/newick_display/1.6+galaxy1 |
| 64 | Group | Grouping1 |
| 65 | Tabular-to-FASTA | toolshed.g2.bx.psu.edu/repos/devteam/tabular_to_fasta/tab2fasta/1.1.1 |
| 66 | Tabular-to-FASTA | toolshed.g2.bx.psu.edu/repos/devteam/tabular_to_fasta/tab2fasta/1.1.1 |
| 67 | FASTA Merge Files and Filter Unique Sequences | toolshed.g2.bx.psu.edu/repos/galaxyp/fasta_merge_files_and_filter_unique_sequences/fasta_merge_files_and_filter_unique_sequences/1.2.0 |
| 68 | FASTA Merge Files and Filter Unique Sequences | toolshed.g2.bx.psu.edu/repos/galaxyp/fasta_merge_files_and_filter_unique_sequences/fasta_merge_files_and_filter_unique_sequences/1.2.0 |
| 69 | ClustalW | toolshed.g2.bx.psu.edu/repos/devteam/clustalw/clustalw/2.1+galaxy1 |
| 70 | ClustalW | toolshed.g2.bx.psu.edu/repos/devteam/clustalw/clustalw/2.1+galaxy1 |
| 71 | FASTTREE | toolshed.g2.bx.psu.edu/repos/iuc/fasttree/fasttree/2.1.10+galaxy1 |
| 72 | FASTTREE | toolshed.g2.bx.psu.edu/repos/iuc/fasttree/fasttree/2.1.10+galaxy1 |
| 73 | Newick Display | toolshed.g2.bx.psu.edu/repos/iuc/newick_utils/newick_display/1.6+galaxy1 |
| 74 | Newick Display | toolshed.g2.bx.psu.edu/repos/iuc/newick_utils/newick_display/1.6+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| number_of_reads_before_host_removal_and_number_of_host_reads_found_per_sample_fig | number_of_reads_before_host_removal_and_number_of_host_reads_found_per_sample_fig | n/a |
|
| removed_host_percentage_fig | removed_host_percentage_fig | n/a |
|
| mapping_mean_depth_per_sample_fig | mapping_mean_depth_per_sample_fig | n/a |
|
| mapping_coverage_percentage_per_sample_fig | mapping_coverage_percentage_per_sample_fig | n/a |
|
| number_of_Variants_and_SNPs_indentified_fig | number_of_Variants_and_SNPs_indentified_fig | n/a |
|
| all_samples_contigs_in_one_fasta_file | all_samples_contigs_in_one_fasta_file | n/a |
|
| all_vfs_in_one_tabular | all_vfs_in_one_tabular | n/a |
|
| split_by_group_collection | split_by_group_collection | n/a |
|
| adjusted_abricate_vfs_tabular_part1 | adjusted_abricate_vfs_tabular_part1 | n/a |
|
| adjusted_abricate_vfs_tabular_part2 | adjusted_abricate_vfs_tabular_part2 | n/a |
|
| amrs_count | amrs_count | n/a |
|
| vfs_count | vfs_count | n/a |
|
| heatmap_table | heatmap_table | n/a |
|
| filtered_sequences_with_vfs_fasta | filtered_sequences_with_vfs_fasta | n/a |
|
| heatmap_pdf | heatmap_pdf | n/a |
|
| heatmap_png | heatmap_png | n/a |
|
| clustalw_on_input_dnd | clustalw_on_input_dnd | n/a |
|
| clustalw_on_input_clustal | clustalw_on_input_clustal | n/a |
|
| vfs_amrs_count_table | vfs_amrs_count_table | n/a |
|
| filtered_empty_datasets | filtered_empty_datasets | n/a |
|
| fasttree_nhx | fasttree_nhx | n/a |
|
| newick_genes_tree_graphs_collection | newick_genes_tree_graphs_collection | n/a |
|
| all_samples_phylogenetic_tree_based_amrs | all_samples_phylogenetic_tree_based_amrs | n/a |
|
| all_samples_phylogenetic_tree_based_vfs | all_samples_phylogenetic_tree_based_vfs | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 235 Downloads: 35 Runs: 0
Created: 2nd Jun 2025 at 10:55
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master