Workflow Type: Galaxy
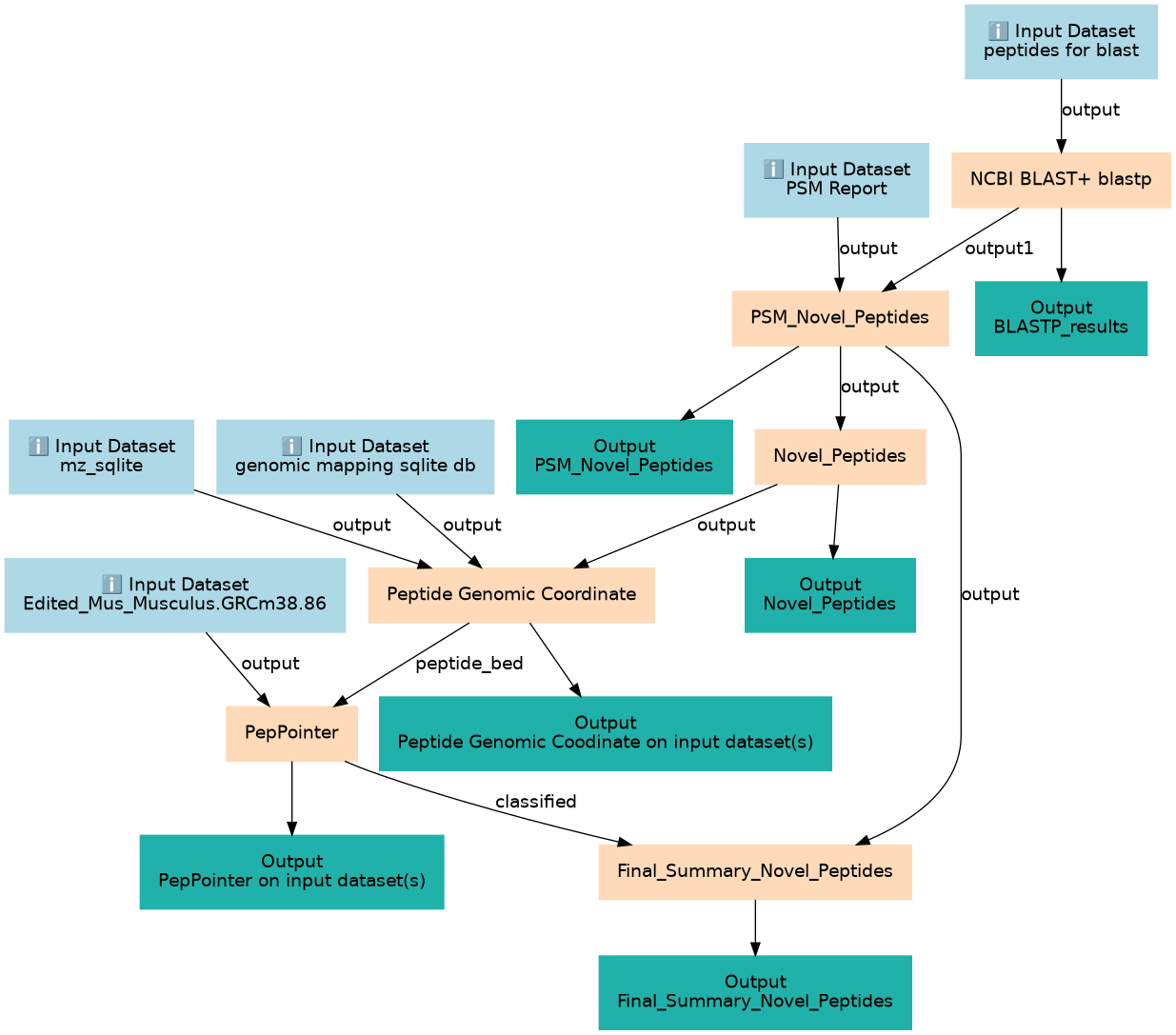
Proteogenomics 3: Novel peptide analysis
Associated Tutorial
This workflows is part of the tutorial Proteogenomics 3: Novel peptide analysis, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Tutorial Author(s): Subina Mehta, Timothy J. Griffin, Pratik Jagtap, Ray Sajulga, James Johnson, Praveen Kumar
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Edited_Mus_Musculus.GRCm38.86 | #main/Edited_Mus_Musculus.GRCm38.86 | n/a |
|
| PSM Report | #main/PSM Report | n/a |
|
| genomic mapping sqlite db | #main/genomic mapping sqlite db | n/a |
|
| mz_sqlite | #main/mz_sqlite | n/a |
|
| peptides for blast | #main/peptides for blast | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 5 | NCBI BLAST+ blastp | toolshed.g2.bx.psu.edu/repos/devteam/ncbi_blast_plus/ncbi_blastp_wrapper/2.16.0+galaxy0 |
| 6 | PSM_Novel_Peptides | toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
| 7 | Novel_Peptides | toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
| 8 | Peptide Genomic Coordinate | toolshed.g2.bx.psu.edu/repos/galaxyp/peptide_genomic_coordinate/peptide_genomic_coordinate/1.0.0 |
| 9 | PepPointer | toolshed.g2.bx.psu.edu/repos/galaxyp/pep_pointer/pep_pointer/0.1.3+galaxy1 |
| 10 | Final_Summary_Novel_Peptides | toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| BLASTP_results | #main/BLASTP_results | n/a |
|
| Final_Summary_Novel_Peptides | #main/Final_Summary_Novel_Peptides | n/a |
|
| Novel_Peptides | #main/Novel_Peptides | n/a |
|
| PSM_Novel_Peptides | #main/PSM_Novel_Peptides | n/a |
|
| PepPointer on input dataset(s) | #main/PepPointer on input dataset(s) | n/a |
|
| Peptide Genomic Coodinate on input dataset(s) | #main/Peptide Genomic Coodinate on input dataset(s) | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tool
Activity
Views: 222 Downloads: 32 Runs: 0
Created: 2nd Jun 2025 at 10:54
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master