Workflow Type: Galaxy
Associated Tutorial
This workflows is part of the tutorial MaxQuant and MSstats for the analysis of label-free data, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Tutorial Author(s): Melanie Föll, Matthias Fahrner
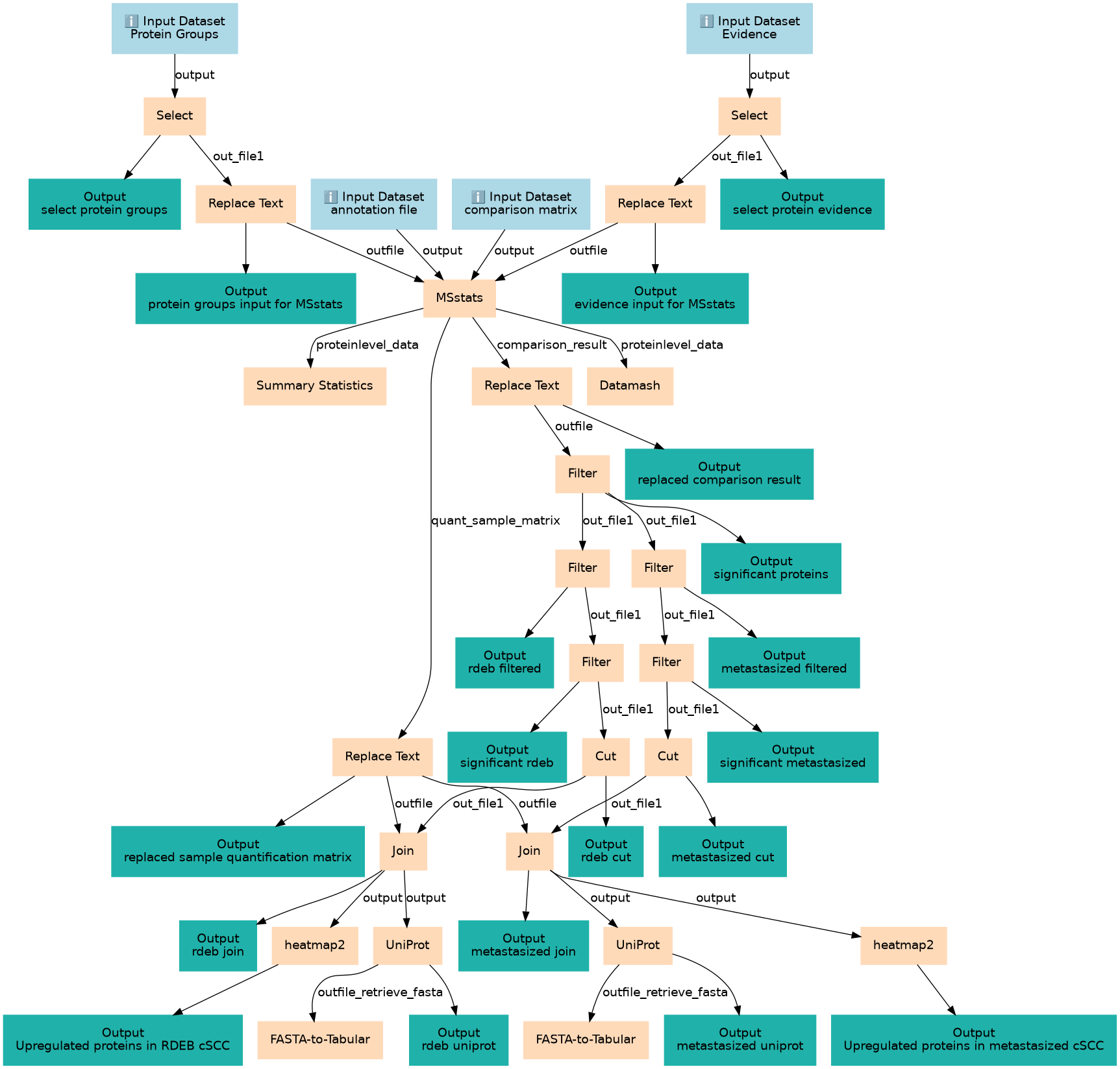
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Evidence | Evidence | n/a |
|
| Protein Groups | Protein Groups | n/a |
|
| annotation file | annotation file | n/a |
|
| comparison matrix | comparison matrix | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Select | Grep1 |
| 5 | Select | Grep1 |
| 6 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.5+galaxy0 |
| 7 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.5+galaxy0 |
| 8 | MSstats | toolshed.g2.bx.psu.edu/repos/galaxyp/msstats/msstats/4.0.0+galaxy1 |
| 9 | Summary Statistics | Summary_Statistics1 |
| 10 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.5+galaxy0 |
| 11 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.5+galaxy0 |
| 12 | Datamash | toolshed.g2.bx.psu.edu/repos/iuc/datamash_ops/datamash_ops/1.8+galaxy0 |
| 13 | Filter | Filter1 |
| 14 | Filter | Filter1 |
| 15 | Filter | Filter1 |
| 16 | Filter | Filter1 |
| 17 | Filter | Filter1 |
| 18 | Cut | Cut1 |
| 19 | Cut | Cut1 |
| 20 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/9.5+galaxy0 |
| 21 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/9.5+galaxy0 |
| 22 | UniProt | toolshed.g2.bx.psu.edu/repos/bgruening/uniprot_rest_interface/uniprot/0.5 |
| 23 | heatmap2 | toolshed.g2.bx.psu.edu/repos/iuc/ggplot2_heatmap2/ggplot2_heatmap2/3.2.0+galaxy1 |
| 24 | heatmap2 | toolshed.g2.bx.psu.edu/repos/iuc/ggplot2_heatmap2/ggplot2_heatmap2/3.2.0+galaxy1 |
| 25 | UniProt | toolshed.g2.bx.psu.edu/repos/bgruening/uniprot_rest_interface/uniprot/0.5 |
| 26 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.1 |
| 27 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| select protein groups | select protein groups | n/a |
|
| select protein evidence | select protein evidence | n/a |
|
| protein groups input for MSstats | protein groups input for MSstats | n/a |
|
| evidence input for MSstats | evidence input for MSstats | n/a |
|
| replaced sample quantification matrix | replaced sample quantification matrix | n/a |
|
| replaced comparison result | replaced comparison result | n/a |
|
| significant proteins | significant proteins | n/a |
|
| metastasized filtered | metastasized filtered | n/a |
|
| rdeb filtered | rdeb filtered | n/a |
|
| significant metastasized | significant metastasized | n/a |
|
| significant rdeb | significant rdeb | n/a |
|
| metastasized cut | metastasized cut | n/a |
|
| rdeb cut | rdeb cut | n/a |
|
| metastasized join | metastasized join | n/a |
|
| rdeb join | rdeb join | n/a |
|
| metastasized uniprot | metastasized uniprot | n/a |
|
| Upregulated proteins in metastasized cSCC | Upregulated proteins in metastasized cSCC | n/a |
|
| Upregulated proteins in RDEB cSCC | Upregulated proteins in RDEB cSCC | n/a |
|
| rdeb uniprot | rdeb uniprot | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 40 Downloads: 4 Runs: 0
Created: 2nd Jun 2025 at 10:52
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master