Workflow Type: Galaxy
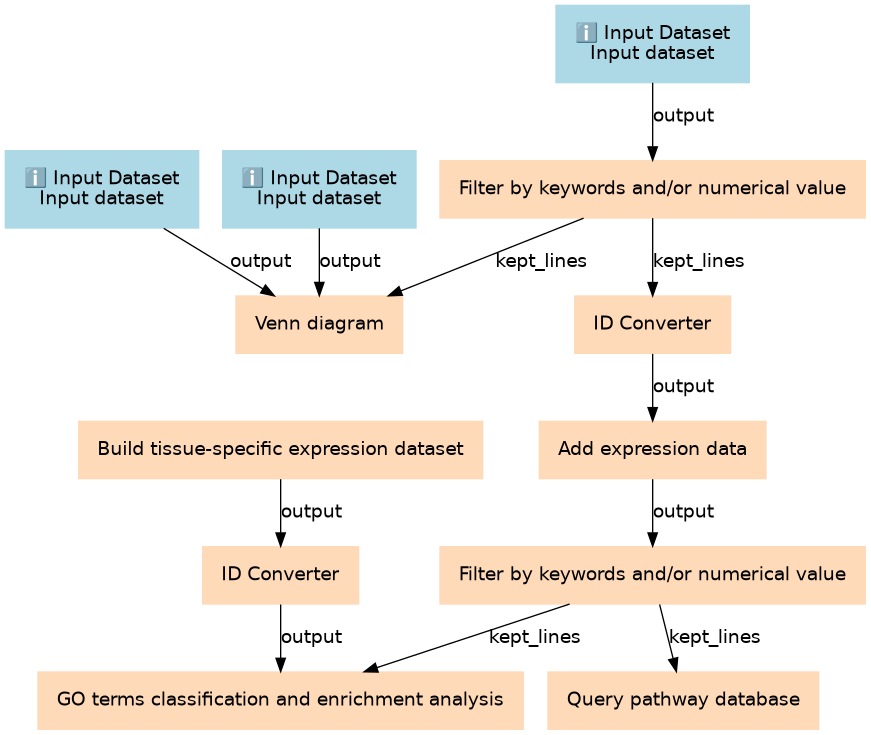
Annotating a protein list identified by LC-MS/MS experiments
Associated Tutorial
This workflows is part of the tutorial Annotating a protein list identified by LC-MS/MS experiments, available in the GTN
Thanks to...
Tutorial Author(s): Valentin Loux, Florence Combes, David Christiany, Yves Vandenbrouck
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Build tissue-specific expression dataset | toolshed.g2.bx.psu.edu/repos/proteore/proteore_tissue_specific_expression_data/retrieve_from_hpa/2019.02.27 |
| 4 | Filter by keywords and/or numerical value | toolshed.g2.bx.psu.edu/repos/proteore/proteore_filter_keywords_values/MQoutputfilter/2019.03.11 |
| 5 | ID Converter | toolshed.g2.bx.psu.edu/repos/proteore/proteore_id_converter/IDconverter/2019.03.07 |
| 6 | ID Converter | toolshed.g2.bx.psu.edu/repos/proteore/proteore_id_converter/IDconverter/2019.03.07 |
| 7 | Venn diagram | toolshed.g2.bx.psu.edu/repos/proteore/proteore_venn_diagram/Jvenn/2019.02.21 |
| 8 | Add expression data | toolshed.g2.bx.psu.edu/repos/proteore/proteore_expression_rnaseq_abbased/rna_abbased_data/2019.03.07 |
| 9 | Filter by keywords and/or numerical value | toolshed.g2.bx.psu.edu/repos/proteore/proteore_filter_keywords_values/MQoutputfilter/2019.03.11 |
| 10 | GO terms classification and enrichment analysis | toolshed.g2.bx.psu.edu/repos/proteore/proteore_clusterprofiler/cluter_profiler/2019.02.18 |
| 11 | Query pathway database | toolshed.g2.bx.psu.edu/repos/proteore/proteore_reactome/reactome_analysis/2019.03.05 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 217 Downloads: 27 Runs: 0
Created: 2nd Jun 2025 at 10:52
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master