Workflow Type: Galaxy
Peptide Library Data Analysis
Associated Tutorial
This workflows is part of the tutorial Peptide Library Data Analysis, available in the GTN
Thanks to...
Tutorial Author(s): Jayadev Joshi, Daniel Blankenberg
Steps
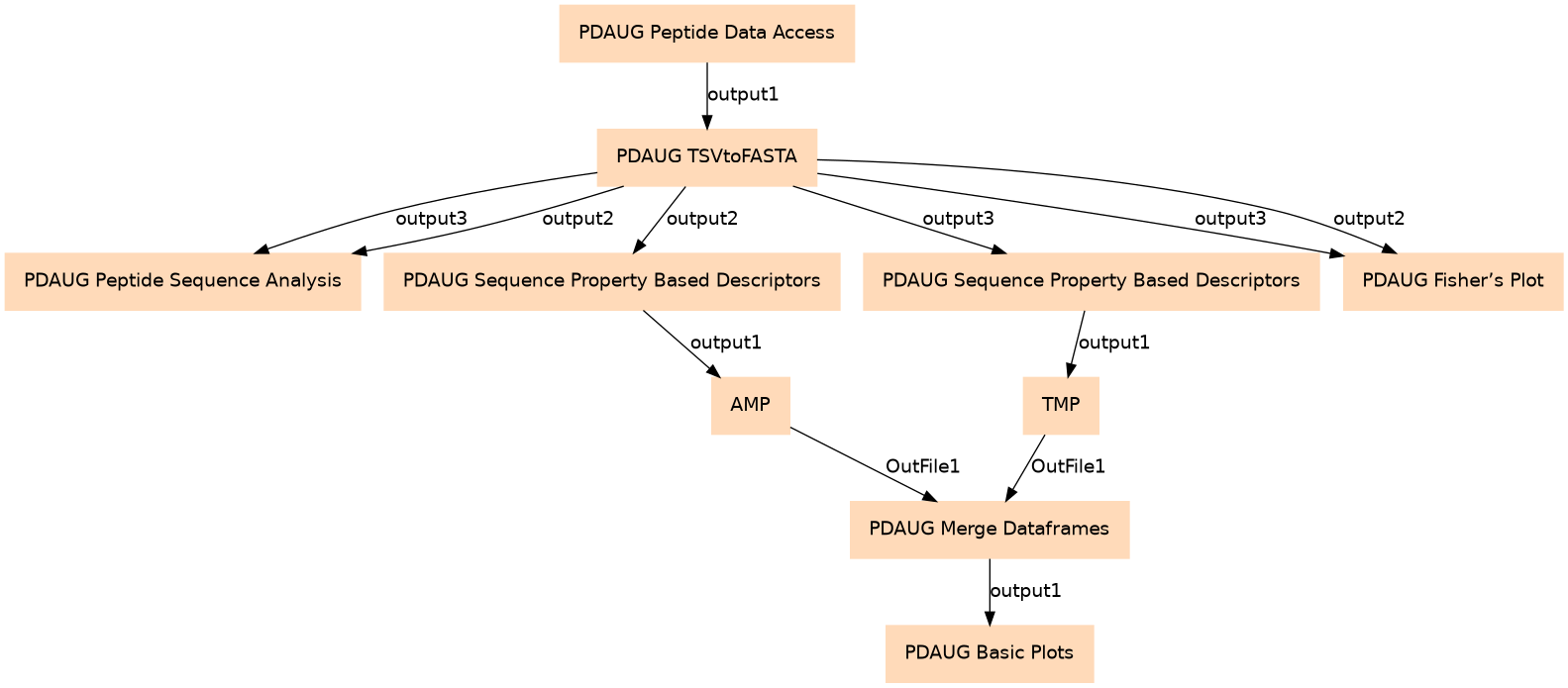
| ID | Name | Description |
|---|---|---|
| 0 | PDAUG Peptide Data Access | toolshed.g2.bx.psu.edu/repos/jay/pdaug_peptide_data_access/pdaug_peptide_data_access/0.1.0 |
| 1 | PDAUG TSVtoFASTA | toolshed.g2.bx.psu.edu/repos/jay/pdaug_tsvtofasta/pdaug_tsvtofasta/0.1.0 |
| 2 | PDAUG Peptide Sequence Analysis | toolshed.g2.bx.psu.edu/repos/jay/pdaug_peptide_sequence_analysis/pdaug_peptide_sequence_analysis/0.1.0 |
| 3 | PDAUG Sequence Property Based Descriptors | toolshed.g2.bx.psu.edu/repos/jay/pdaug_sequence_property_based_descriptors/pdaug_sequence_property_based_descriptors/0.1.0 |
| 4 | PDAUG Sequence Property Based Descriptors | toolshed.g2.bx.psu.edu/repos/jay/pdaug_sequence_property_based_descriptors/pdaug_sequence_property_based_descriptors/0.1.0 |
| 5 | PDAUG Fisher's Plot | toolshed.g2.bx.psu.edu/repos/jay/pdaug_fishers_plot/pdaug_fishers_plot/0.1.0 |
| 6 | AMP | toolshed.g2.bx.psu.edu/repos/jay/pdaug_addclasslabel/pdaug_addclasslabel/0.1.0 |
| 7 | TMP | toolshed.g2.bx.psu.edu/repos/jay/pdaug_addclasslabel/pdaug_addclasslabel/0.1.0 |
| 8 | PDAUG Merge Dataframes | toolshed.g2.bx.psu.edu/repos/jay/pdaug_merge_dataframes/pdaug_merge_dataframes/0.1.0 |
| 9 | PDAUG Basic Plots | toolshed.g2.bx.psu.edu/repos/jay/pdaug_basic_plots/pdaug_basic_plots/0.1.0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 240 Downloads: 42 Runs: 0
Created: 2nd Jun 2025 at 10:51
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master