Workflow Type: Galaxy
Frozen
This is part of a series of workflows to annotate a genome, tagged with TSI-annotation.
These workflows are based on command-line code by Luke Silver, converted into Galaxy Australia workflows.
The workflows can be run in this order:
- Repeat masking
- RNAseq QC and read trimming
- Find transcripts
- Combine transcripts
- Extract transcripts
- Convert formats
- Fgenesh annotation
About this workflow:
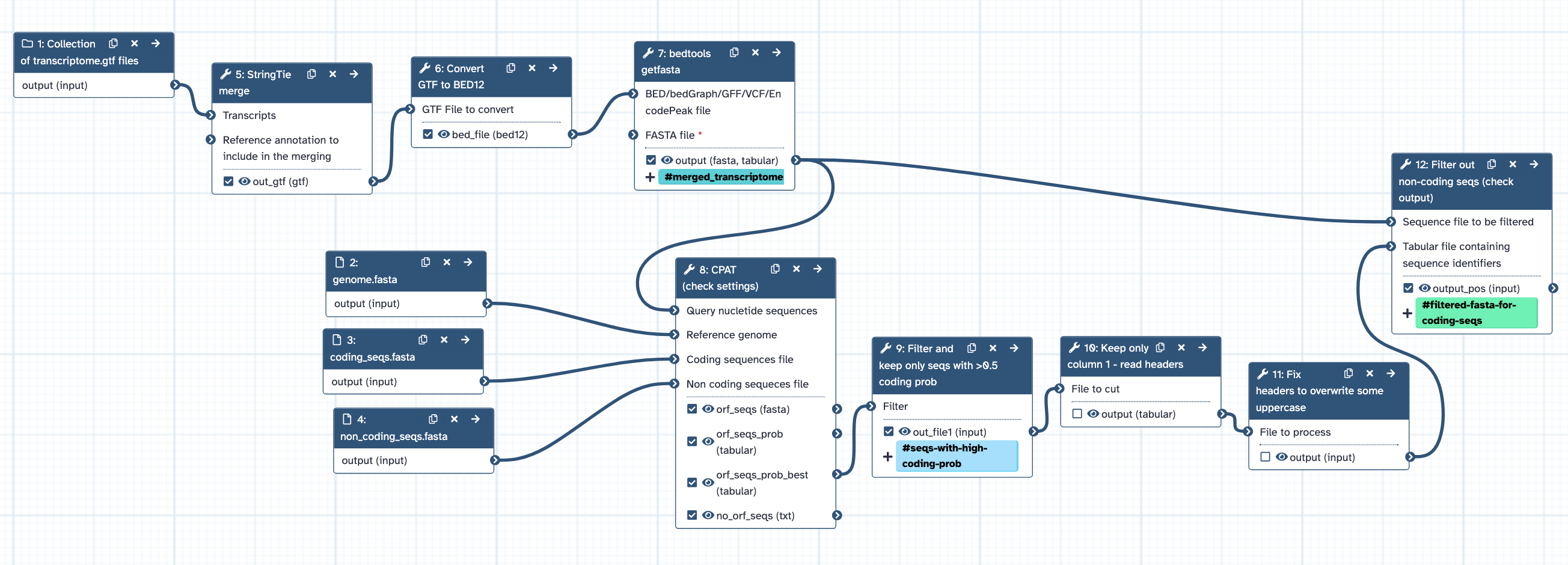
- Inputs: multiple transcriptome.gtfs from different tissues, genome.fasta, coding_seqs.fasta, non_coding_seqs.fasta
- Runs StringTie merge to combine transcriptomes, with default settings except for -m = 30 and -F = 0.1, to produce a merged_transcriptomes.gtf.
- Runs Convert GTF to BED12 with default settings, to produce a merged_transcriptomes.bed.
- Runs bedtools getfasta with default settings except for -name = yes, -s = yes, -split - yes, to produce a merged_transcriptomes.fasta
- Runs CPAT to generate seqs with high coding probability.
- Filters out non-coding seqs from the merged_transcriptomes.fasta
- Output: filtered_merged_transcriptomes.fasta
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Collection of transcriptome.gtf files | #main/Collection of transcriptome.gtf files | n/a |
|
| coding_seqs.fasta | #main/coding_seqs.fasta | n/a |
|
| genome.fasta | #main/genome.fasta | n/a |
|
| non_coding_seqs.fasta | #main/non_coding_seqs.fasta | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | StringTie merge | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie_merge/2.2.1+galaxy1 |
| 5 | Convert GTF to BED12 | toolshed.g2.bx.psu.edu/repos/iuc/gtftobed12/gtftobed12/357 |
| 6 | bedtools getfasta | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_getfastabed/2.30.0+galaxy1 |
| 7 | CPAT (check settings) | The table of best probabilities is called orf_seqs_prob_best; converted this to tabular toolshed.g2.bx.psu.edu/repos/bgruening/cpat/cpat/3.0.5+galaxy0 |
| 8 | Filter and keep only seqs with >0.5 coding prob | skipping 1 header line Filter1 |
| 9 | Keep only column 1 - read headers | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/9.3+galaxy0 |
| 10 | Fix headers to overwrite some uppercase | part of the headers have become capitalized, this reverts everything after the :: to lowercase. May need to be changed if headers don't have the same format with a :: in them. toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sed_tool/9.3+galaxy0 |
| 11 | Filter out non-coding seqs (check output) | toolshed.g2.bx.psu.edu/repos/peterjc/seq_filter_by_id/seq_filter_by_id/0.2.9 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| bed_file | #main/bed_file | n/a |
|
| no_orf_seqs | #main/no_orf_seqs | n/a |
|
| orf_seqs | #main/orf_seqs | n/a |
|
| orf_seqs_prob | #main/orf_seqs_prob | n/a |
|
| orf_seqs_prob_best | #main/orf_seqs_prob_best | n/a |
|
| out_file1 | #main/out_file1 | n/a |
|
| out_gtf | #main/out_gtf | n/a |
|
| output | #main/output | n/a |
|
| output_pos | #main/output_pos | n/a |
|
Version History
Version 1 (earliest) Created 8th May 2024 at 08:07 by Anna Syme
Initial commit
Frozen
 Version-1
Version-1ff43cfe
 Creators and Submitter
Creators and SubmitterCreators
Submitter
Citation
Silver, L., & Syme, A. (2024). Combine transcripts - TSI. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.878.1
Activity
Views: 3373 Downloads: 351 Runs: 0
Created: 8th May 2024 at 08:07
Last updated: 9th May 2024 at 05:06
 Tags
Tags Attributions
AttributionsNone
 Collections
Collections Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673