Workflow Type: Galaxy
Open
Stable
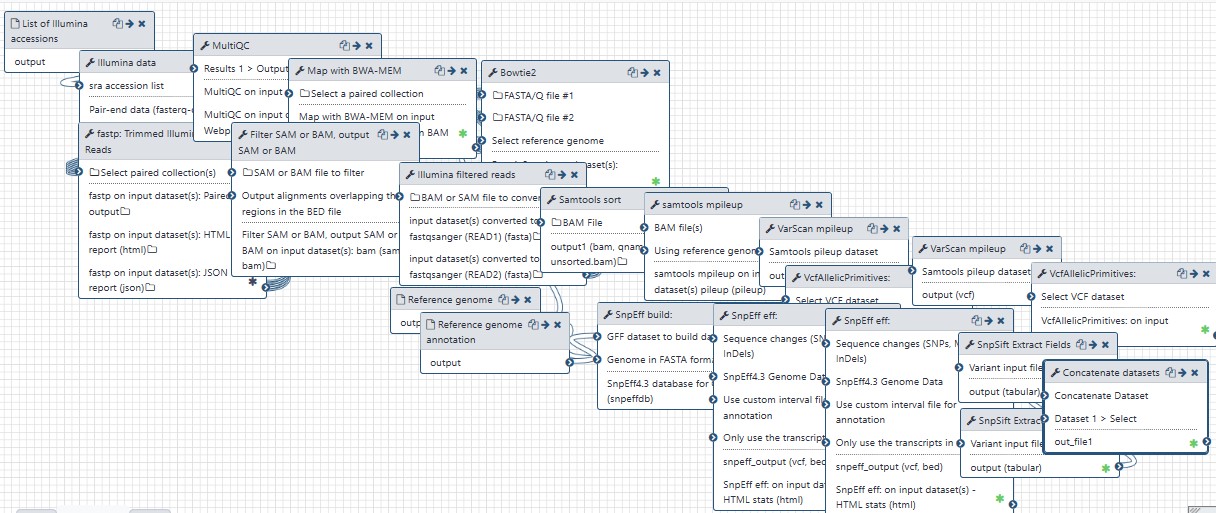
Detects SNPs and INDELs using VARSCAN2.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| List of Illumina accessions | List of Illumina accessions | n/a |
|
| Reference genome | Reference genome | n/a |
|
| Reference genome annotation | Reference genome annotation | COVID-19 |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Illumina data | toolshed.g2.bx.psu.edu/repos/iuc/sra_tools/fasterq_dump/2.10.4 |
| 4 | SnpEff build: | toolshed.g2.bx.psu.edu/repos/iuc/snpeff/snpEff_build_gb/4.3+T.galaxy4 |
| 5 | fastp: Trimmed Illumina Reads | toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.19.5+galaxy1 |
| 6 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.7 |
| 7 | Map with BWA-MEM | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.17.1 |
| 8 | Filter SAM or BAM, output SAM or BAM | toolshed.g2.bx.psu.edu/repos/devteam/samtool_filter2/samtool_filter2/1.8+galaxy1 |
| 9 | Illumina filtered reads | toolshed.g2.bx.psu.edu/repos/iuc/samtools_fastx/samtools_fastx/1.9+galaxy1 |
| 10 | Bowtie2 | toolshed.g2.bx.psu.edu/repos/devteam/bowtie2/bowtie2/2.3.4.3+galaxy0 |
| 11 | Samtools sort | toolshed.g2.bx.psu.edu/repos/devteam/samtools_sort/samtools_sort/2.0.3 |
| 12 | samtools mpileup | toolshed.g2.bx.psu.edu/repos/devteam/samtools_mpileup/samtools_mpileup/2.1.4 |
| 13 | VarScan mpileup | toolshed.g2.bx.psu.edu/repos/iuc/varscan_mpileup/varscan_mpileup/2.4.3.1 |
| 14 | VarScan mpileup | toolshed.g2.bx.psu.edu/repos/iuc/varscan_mpileup/varscan_mpileup/2.4.3.1 |
| 15 | VcfAllelicPrimitives: | toolshed.g2.bx.psu.edu/repos/devteam/vcfallelicprimitives/vcfallelicprimitives/1.0.0_rc3+galaxy0 |
| 16 | VcfAllelicPrimitives: | toolshed.g2.bx.psu.edu/repos/devteam/vcfallelicprimitives/vcfallelicprimitives/1.0.0_rc3+galaxy0 |
| 17 | SnpEff eff: | toolshed.g2.bx.psu.edu/repos/iuc/snpeff/snpEff/4.3+T.galaxy1 |
| 18 | SnpEff eff: | toolshed.g2.bx.psu.edu/repos/iuc/snpeff/snpEff/4.3+T.galaxy1 |
| 19 | SnpSift Extract Fields | toolshed.g2.bx.psu.edu/repos/iuc/snpsift/snpSift_extractFields/4.3+t.galaxy0 |
| 20 | SnpSift Extract Fields | toolshed.g2.bx.psu.edu/repos/iuc/snpsift/snpSift_extractFields/4.3+t.galaxy0 |
| 21 | Concatenate datasets | cat1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_4 | _anonymous_output_4 | n/a |
|
| _anonymous_output_5 | _anonymous_output_5 | n/a |
|
| _anonymous_output_6 | _anonymous_output_6 | n/a |
|
| _anonymous_output_7 | _anonymous_output_7 | n/a |
|
| _anonymous_output_8 | _anonymous_output_8 | n/a |
|
| _anonymous_output_9 | _anonymous_output_9 | n/a |
|
| _anonymous_output_10 | _anonymous_output_10 | n/a |
|
| _anonymous_output_11 | _anonymous_output_11 | n/a |
|
| _anonymous_output_12 | _anonymous_output_12 | n/a |
|
| _anonymous_output_13 | _anonymous_output_13 | n/a |
|
| _anonymous_output_14 | _anonymous_output_14 | n/a |
|
| _anonymous_output_15 | _anonymous_output_15 | n/a |
|
| _anonymous_output_16 | _anonymous_output_16 | n/a |
|
| _anonymous_output_17 | _anonymous_output_17 | n/a |
|
| _anonymous_output_18 | _anonymous_output_18 | n/a |
|
| _anonymous_output_19 | _anonymous_output_19 | n/a |
|
| _anonymous_output_20 | _anonymous_output_20 | n/a |
|
| _anonymous_output_21 | _anonymous_output_21 | n/a |
|
| _anonymous_output_22 | _anonymous_output_22 | n/a |
|
| _anonymous_output_23 | _anonymous_output_23 | n/a |
|
| _anonymous_output_24 | _anonymous_output_24 | n/a |
|
| _anonymous_output_25 | _anonymous_output_25 | n/a |
|
| _anonymous_output_26 | _anonymous_output_26 | n/a |
|
| _anonymous_output_27 | _anonymous_output_27 | n/a |
|
| _anonymous_output_28 | _anonymous_output_28 | n/a |
|
| _anonymous_output_29 | _anonymous_output_29 | n/a |
|
Version History
Version 1 (earliest) Created 18th Jun 2020 at 23:57 by Ambarish Kumar
Added/updated 2 files
Open
 master
masterde69d64
 Visit source
Visit source Run on Galaxy
Run on Galaxy Creators and Submitter
Creators and Submitter