Workflow Type: Galaxy
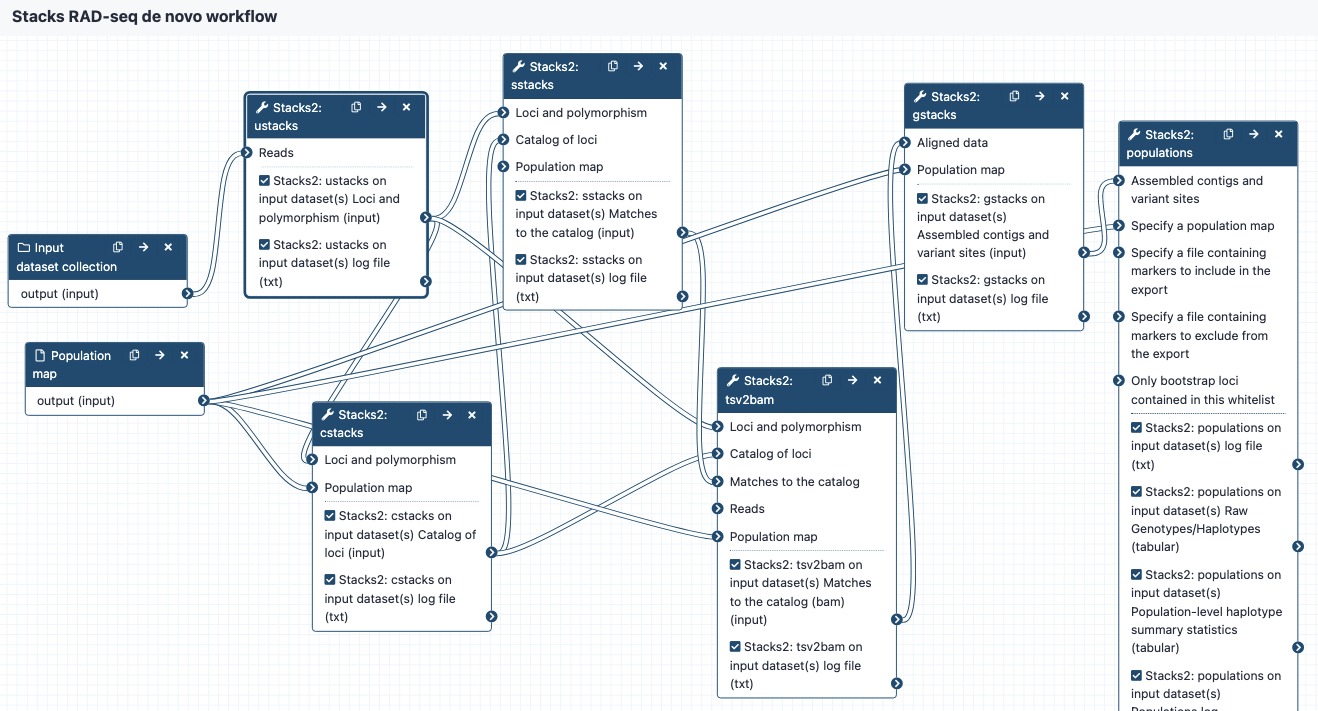
workflow-denovo-stacks
These workflows are part of a set designed to work for RAD-seq data on the Galaxy platform, using the tools from the Stacks program.
Galaxy Australia: https://usegalaxy.org.au/
Stacks: http://catchenlab.life.illinois.edu/stacks/
Inputs
- demultiplexed reads in fastq format, may be output from the QC workflow. Files are in a collection.
- population map in text format
Steps and outputs
ustacks:
- input reads go to ustacks.
- ustacks assembles the reads into matching stacks (hypothetical alleles).
- The outputs are in a collection called something like: Stacks2: ustacks on data 21, data 20, and others Loci and polymorphism. Click on this to see the files:
- for each sample, assembled loci (tsv format), named e.g. sample_CAAC.tags
- for each sample, model calls from each locus (tsv format), named e.g. sample_CAAC.snps
- for each sample, haplotypes/alleles recorded from each locus (tsv format), named e.g. sample_CAAC.alleles
- Please see sections 6.1 to 6.4 in https://catchenlab.life.illinois.edu/stacks/manual/#ufiles for a full description.
cstacks:
- cstacks will merge stacks into a catalog of consensus loci.
- The outputs are in a collection called something like Stacks2: cstacks on data 3, data 71, and others Catalog of loci. Click on this to see the three files, each in tsv format: catalog.tags catalog.snps catalog.alleles
sstacks:
- sstacks will compare each sample to the loci in the catalog.
- The outputs are in a collection called something like Stacks2: sstacks on data 3, data 76, and others Matches to the catalog.Click on this to see the files: There is one file for each sample, named e.g. sample_CAAC.matches, in tsv format.
tsv2bam:
- Conversion to BAM format
- Reads from each sample are now aligned to each locus, and the tsv2bam tool will convert this into a bam file for each sample.
- The outputs are in a collection called something like Stacks2: tsv2bam on data 3, data 94, and others Matches to the catalog.Click on this to see the files: There is one file for each sample, named e.g sample_CAAC.matches, in BAM format.
gstacks:
- Catalog of loci in fasta format
- Variant calls in VCF format
populations:
- Locus consensus sequences in fasta format
- Snp calls, in VCF format
- Haplotypes, in VCF format
- Summary statistics
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Stacks2: ustacks | toolshed.g2.bx.psu.edu/repos/iuc/stacks2_ustacks/stacks2_ustacks/2.55+galaxy3 |
| 3 | Stacks2: cstacks | toolshed.g2.bx.psu.edu/repos/iuc/stacks2_cstacks/stacks2_cstacks/2.55+galaxy2 |
| 4 | Stacks2: sstacks | toolshed.g2.bx.psu.edu/repos/iuc/stacks2_sstacks/stacks2_sstacks/2.55+galaxy2 |
| 5 | Stacks2: tsv2bam | toolshed.g2.bx.psu.edu/repos/iuc/stacks2_tsv2bam/stacks2_tsv2bam/2.55+galaxy2 |
| 6 | Stacks2: gstacks | toolshed.g2.bx.psu.edu/repos/iuc/stacks2_gstacks/stacks2_gstacks/2.55+galaxy2 |
| 7 | Stacks2: populations | toolshed.g2.bx.psu.edu/repos/iuc/stacks2_populations/stacks2_populations/2.55+galaxy2 |
Version History
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Tool
License
Activity
Views: 6398 Downloads: 431 Runs: 1
Created: 31st May 2022 at 08:39
Annotated Properties
 Tags
TagsThis item has not yet been tagged.
 Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub Run on Galaxy
Run on Galaxy v1.0
v1.0 https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673