Workflow Type: Galaxy
workflow-qc-of-radseq-reads
These workflows are part of a set designed to work for RAD-seq data on the Galaxy platform, using the tools from the Stacks program.
Galaxy Australia: https://usegalaxy.org.au/
Stacks: http://catchenlab.life.illinois.edu/stacks/
Inputs
- demultiplexed reads in fastq format, in a collection
- two adapter sequences in fasta format, for input into cutadapt
Steps and outputs
The workflow can be modified to suit your own parameters.
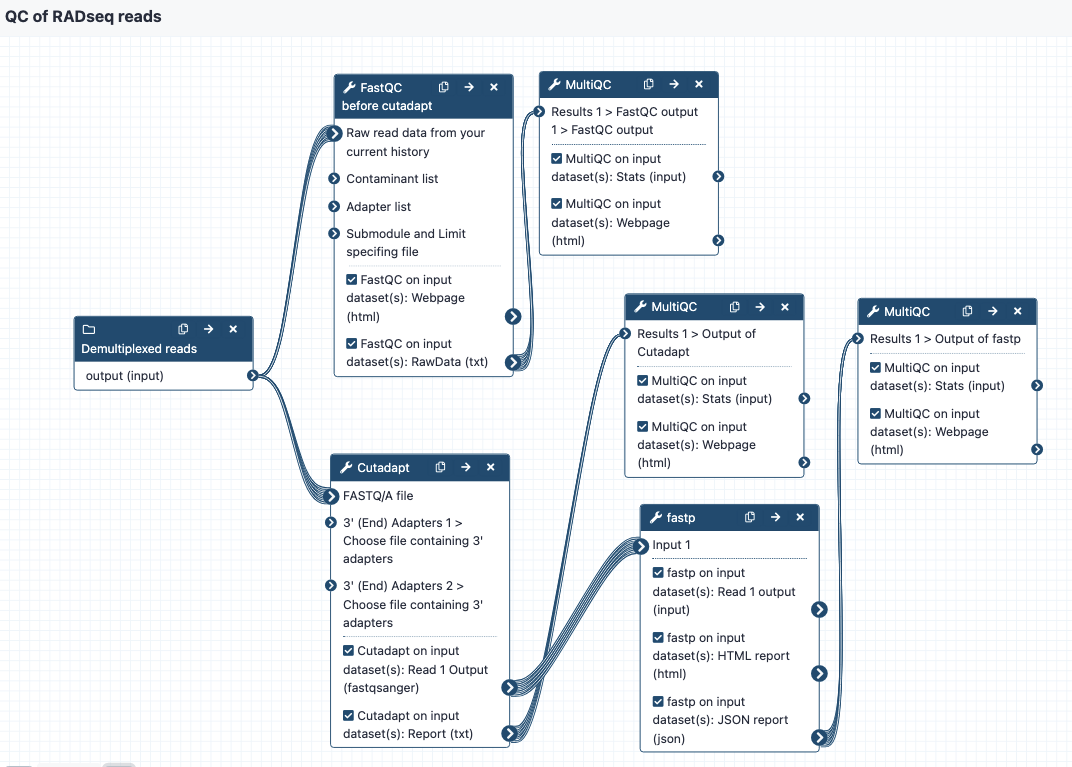
The workflow steps are:
- Run FastQC to get statistics on the raw reads, send to MultiQC to create a nice output. This is tagged as "Report 1" in the Galaxy history.
- Run Cutadapt on the reads to cut adapters - enter two files with adapter sequence at the workflow option for "Choose file containing 3' adapters". The default settings are on except that the "Maximum error rate" for the adapters is set to 0.2 instead of 0.1. Send output statistics to MulitQC, this is "Report 2" in the Galaxy history. Note that you may have different requirements here in terms of how many adapter sequences you want to enter. We recommend copying the workflow and modifying as needed.
- Send these reads to fastp for additional filtering or trimming. Default settings are on but can be modified as needed. Send output statistics to MultiQC, this is "Report 3" in the Galaxy history.
- The filtered and trimmed reads are then ready for the stacks workflows.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Demultiplexed reads | Demultiplexed reads | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | FastQC before cutadapt | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.73+galaxy0 |
| 2 | Cutadapt | Remove adapters from reads. Supply two adapter.fasta files (ask sequencing provider for adapter seqs). Default settings on except max error rate changed to 0.2, and min read length set to 70 toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/4.0+galaxy0 |
| 3 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy0 |
| 4 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy0 |
| 5 | fastp | Additional filtering and trimming options. All defaults are on. toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.23.2+galaxy0 |
| 6 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| FastQC on input dataset(s): Webpage | FastQC on input dataset(s): Webpage | n/a |
|
| FastQC on input dataset(s): RawData | FastQC on input dataset(s): RawData | n/a |
|
| Cutadapt on input dataset(s): Read 1 Output | Cutadapt on input dataset(s): Read 1 Output | n/a |
|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
| MultiQC on input dataset(s): Webpage | MultiQC on input dataset(s): Webpage | n/a |
|
| MultiQC on input dataset(s): Stats | MultiQC on input dataset(s): Stats | n/a |
|
| _anonymous_output_2 | _anonymous_output_2 | n/a |
|
| _anonymous_output_3 | _anonymous_output_3 | n/a |
|
| fastp on input dataset(s): Read 1 output | fastp on input dataset(s): Read 1 output | n/a |
|
| fastp on input dataset(s): HTML report | fastp on input dataset(s): HTML report | n/a |
|
| _anonymous_output_4 | _anonymous_output_4 | n/a |
|
| _anonymous_output_5 | _anonymous_output_5 | n/a |
|
| _anonymous_output_6 | _anonymous_output_6 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreator
Submitter
License
Activity
Views: 5843 Downloads: 396 Runs: 1
Created: 31st May 2022 at 07:55
Annotated Properties
 Tags
TagsThis item has not yet been tagged.
 Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub Run on Galaxy
Run on Galaxy v1.0
v1.0 https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673