Workflow Type: Galaxy
Open
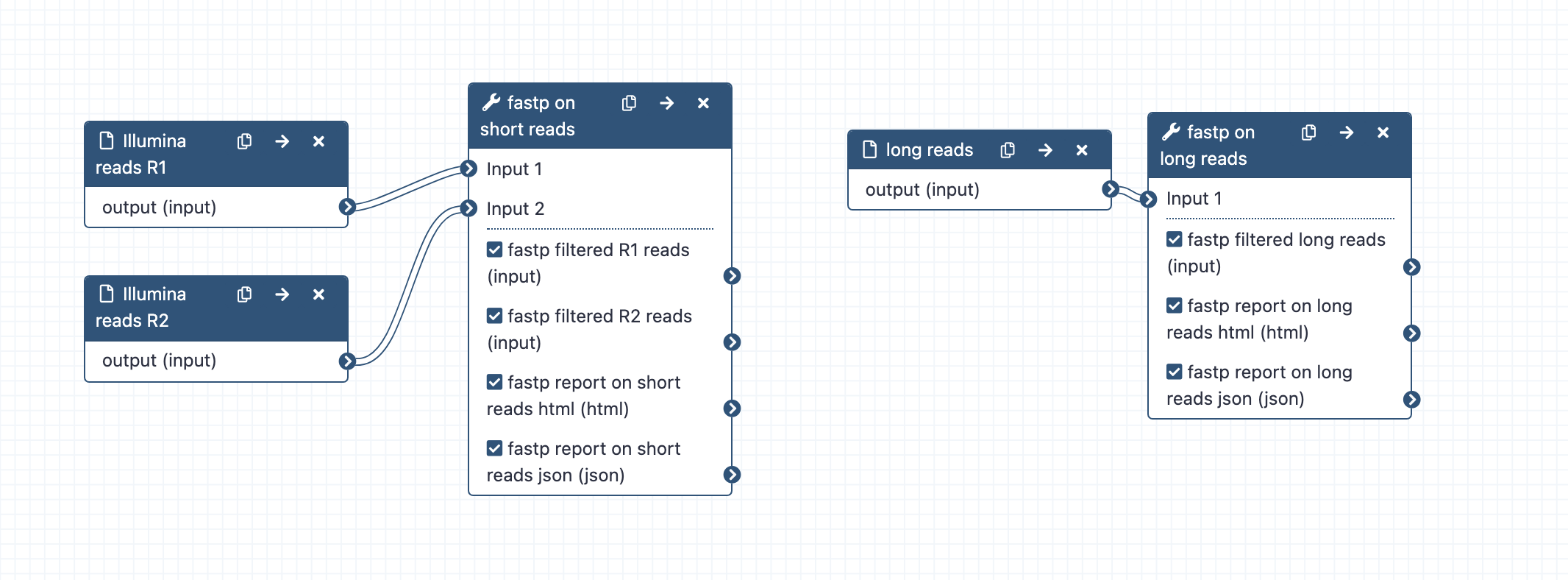
Trim and filter reads; can run alone or as part of a combined workflow for large genome assembly.
- What it does: Trims and filters raw sequence reads according to specified settings.
- Inputs: Long reads (format fastq); Short reads R1 and R2 (format fastq)
- Outputs: Trimmed and filtered reads: fastp_filtered_long_reads.fastq.gz (But note: no trimming or filtering is on by default), fastp_filtered_R1.fastq.gz, fastp_filtered_R2.fastq.gz
- Reports: fastp report on long reads, html; fastp report on short reads, html
- Tools used: fastp (Note. The latest version (0.20.1) of fastp has an issue displaying plot results. Using version 0.19.5 here instead until this is rectified).
- Input parameters: None required, but recommend removing the long reads from the workflow if not using any trimming/filtering settings.
Workflow steps:
Long reads: fastp settings:
- These settings have been changed from the defaults (so that all filtering and trimming settings are now disabled).
- Adapter trimming options: Disable adapter trimming: yes
- Filter options: Quality filtering options: Disable quality filtering: yes
- Filter options: Length filtering options: Disable length filtering: yes
- Read modification options: PolyG tail trimming: Disable
- Output options: output JSON report: yes
Short reads: fastp settings:
- adapter trimming (default setting: adapters are auto-detected)
- quality filtering (default: phred quality 15), unqualified bases limit (default = 40%), number of Ns allowed in a read (default = 5)
- length filtering (default length = min 15)
- polyG tail trimming (default = on for NextSeq/NovaSeq data which is auto detected)
- Output options: output JSON report: yes
Options:
- Change any settings in fastp for any of the input reads.
- Adapter trimming: input the actual adapter sequences. (Alternative tool for long read adapter trimming: Porechop.)
- Trimming n bases from ends of reads if quality less than value x (Alternative tool for trimming long reads: NanoFilt.)
- Discard post-trimmed reads if length is < x (e.g. for long reads, 1000 bp)
- Example filtering/trimming that you might do on long reads: remove adapters (can also be done with Porechop), trim bases from ends of the reads with low quality (can also be done with NanoFilt), after this can keep only reads of length x (e.g. 1000 bp)
Infrastructure_deployment_metadata: Galaxy Australia (Galaxy)
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Illumina reads R1 | Illumina reads R1 | n/a |
|
| Illumina reads R2 | Illumina reads R2 | n/a |
|
| long reads | long reads | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | fastp on short reads | toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.19.5+galaxy1 |
| 4 | fastp on long reads | toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.19.5+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| fastp filtered R2 reads | fastp filtered R2 reads | n/a |
|
| fastp report on short reads html | fastp report on short reads html | n/a |
|
| fastp filtered R1 reads | fastp filtered R1 reads | n/a |
|
| fastp report on short reads json | fastp report on short reads json | n/a |
|
| fastp report on long reads html | fastp report on long reads html | n/a |
|
| fastp filtered long reads | fastp filtered long reads | n/a |
|
| fastp report on long reads json | fastp report on long reads json | n/a |
|
Version History
Version 1 (earliest) Created 8th Nov 2021 at 04:56 by Anna Syme
Added/updated 2 files
Open
 master
master88e7c49
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Tool
Citation
Syme, A. (2021). Trim and filter reads - fastp. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.224.1
License
Activity
Views: 10747 Downloads: 500 Runs: 0
Created: 8th Nov 2021 at 04:56
Last updated: 9th Nov 2021 at 01:11
Annotated Properties
 Attributions
AttributionsNone
 Collections
Collections Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673