Workflow Type: Galaxy
Open
Frozen
Frozen
Frozen
Frozen
Frozen
Frozen
Stable
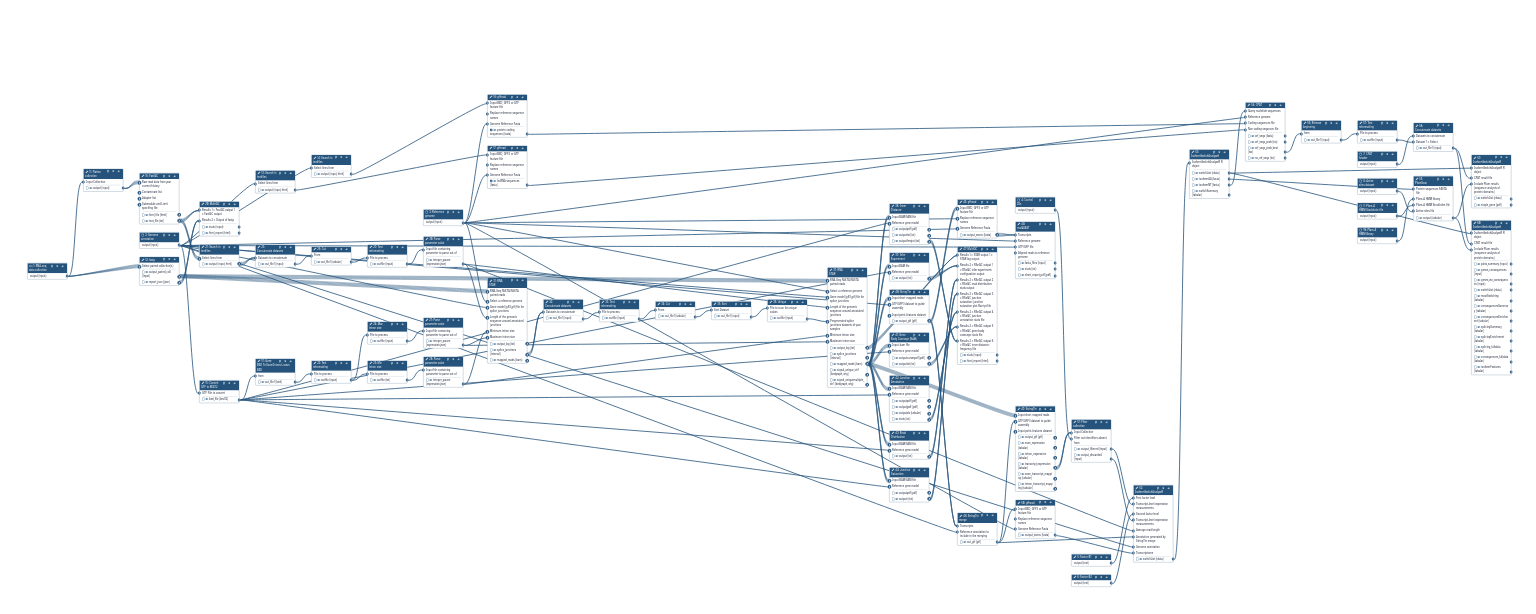
Genome-wide alternative splicing analysis v.2
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Active sites dataset | n/a | Active sites dataset. |
|
| CPAT header | n/a | Galaxy compatible CPAT header. |
|
| Control IDs | n/a | IDs of control sequences, corresponding to filenames. |
|
| Genome annotation | n/a | Reference genome annotation in GTF format. |
|
| Pfam-A HMM Stockholm file | n/a | Stockholm file. |
|
| Pfam-A HMM library | n/a | HMM library. |
|
| RNA-seq data collection | n/a | Collection of paired-end RNA-seq datasets. |
|
| Reference genome | n/a | Genome reference in FASTA format. |
|
Steps
| ID | Name | Description |
|---|---|---|
| 10_Flatten collection | n/a | n/a |
| 11_fastp | n/a | n/a |
| 12_Search in textfiles | n/a | n/a |
| 13_Search in textfiles | n/a | n/a |
| 14_Convert GTF to BED12 | n/a | n/a |
| 15_FastQC | n/a | n/a |
| 16_gffread | n/a | n/a |
| 17_gffread | n/a | n/a |
| 18_Gene BED To Exon_Intron_Codon BED | n/a | n/a |
| 19_MultiQC | n/a | n/a |
| 20_Search in textfiles | n/a | n/a |
| 21_Text reformatting | n/a | n/a |
| 22_Concatenate datasets | n/a | n/a |
| 23_Text reformatting | n/a | n/a |
| 24_Text reformatting | n/a | n/a |
| 25_Cut | n/a | n/a |
| 26_Parse parameter value | n/a | n/a |
| 27_Parse parameter value | n/a | n/a |
| 28_Text reformatting | n/a | n/a |
| 29_Parse parameter value | n/a | n/a |
| 30_RNA STAR | n/a | n/a |
| 31_Concatenate datasets | n/a | n/a |
| 32_Text reformatting | n/a | n/a |
| 33_Cut | n/a | n/a |
| 34_Sort | n/a | n/a |
| 35_Unique | n/a | n/a |
| 36_RNA STAR | n/a | n/a |
| 37_Inner Distance | n/a | n/a |
| 38_Infer Experiment | n/a | n/a |
| 39_StringTie | n/a | n/a |
| 40_Gene Body Coverage (BAM) | n/a | n/a |
| 41_Junction Annotation | n/a | n/a |
| 42_Read Distribution | n/a | n/a |
| 43_Junction Saturation | n/a | n/a |
| 44_gffread | n/a | n/a |
| 45_StringTie merge | n/a | n/a |
| 46_MultiQC | n/a | n/a |
| 47_rnaQUAST | n/a | n/a |
| 48_StringTie | n/a | n/a |
| 49_gffread | n/a | n/a |
| 4_Input parameter | n/a | n/a |
| 50_Filter collection | n/a | n/a |
| 51_IsoformSwitchAnalyzeR | n/a | n/a |
| 52_IsoformSwitchAnalyzeR | n/a | n/a |
| 53_CPAT | n/a | n/a |
| 54_PfamScan | n/a | n/a |
| 55_Remove beginning | n/a | n/a |
| 56_Text reformatting | n/a | n/a |
| 57_Concatenate datasets | n/a | n/a |
| 58_IsoformSwitchAnalyzeR | n/a | n/a |
| 59_IsoformSwitchAnalyzeR | n/a | n/a |
| 5_Input parameter | n/a | n/a |
Version History
Version 7 (latest) Created 11th Jun 2023 at 20:17 by Cristóbal Gallardo
No revision comments
Open
 master
master0f0e4d1
Version 6 Created 11th Jun 2023 at 13:35 by Cristóbal Gallardo
No revision comments
Frozen
 Version-6
Version-67ca27b3
Version 5 Created 11th Jun 2023 at 13:01 by Cristóbal Gallardo
No revision comments
Frozen
 Version-5
Version-5a704afc
Version 4 Created 11th Jun 2023 at 12:57 by Cristóbal Gallardo
No revision comments
Frozen
 Version-4
Version-4820ad68
Version 3 Created 11th Jun 2023 at 12:53 by Cristóbal Gallardo
No revision comments
Frozen
 Version-3
Version-372a6796
Version 2 Created 7th Jun 2023 at 17:10 by Cristóbal Gallardo
No revision comments
Frozen
 Version-2
Version-2c6f2e2d
Version 1 (earliest) Created 25th May 2023 at 23:01 by Cristóbal Gallardo
Initial commit
Frozen
 Version-1
Version-1746fae8
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Citation
Gallardo, C. (2023). Genome-wide alternative splicing analysis v.2. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.482.1
Activity
Views: 9889 Downloads: 2268 Runs: 3
Created: 25th May 2023 at 23:01
Last updated: 7th Jun 2023 at 17:00
Annotated Properties
 Attributions
AttributionsNone
 Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-5752-2155
https://orcid.org/0000-0002-5752-2155