Workflows
What is a Workflow?Filters
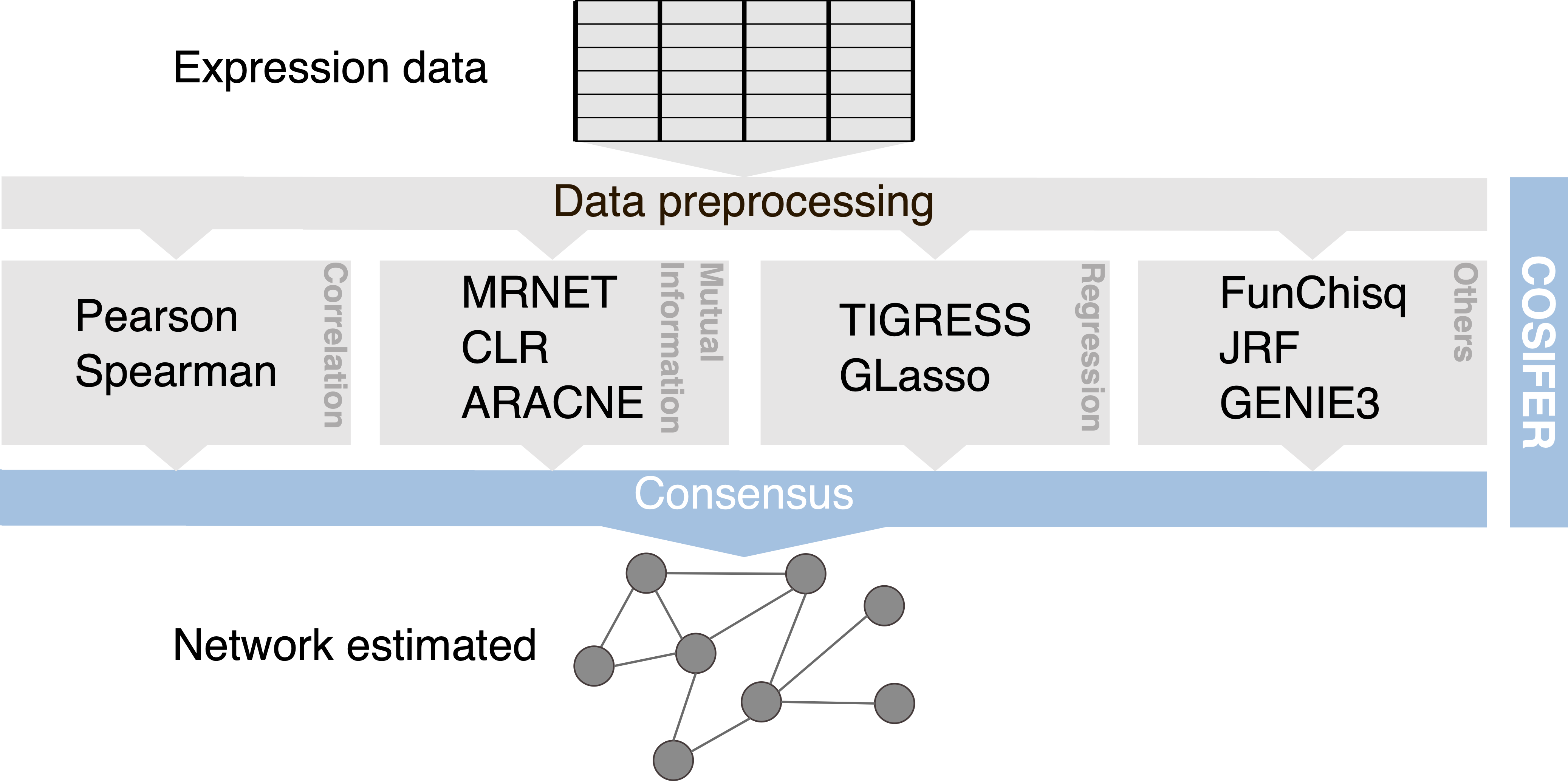
COnSensus Interaction Network InFErence Service
Inference framework for reconstructing networks using a consensus approach between multiple methods and data sources.
Reference
[Manica, Matteo, Charlotte, Bunne, Roland, Mathis, Joris, Cadow, Mehmet Eren, Ahsen, Gustavo A, Stolovitzky, and María Rodríguez, Martínez. "COSIFER: a python package for the consensus inference of molecular ...
VIRify
VIRify is a recently developed pipeline for the detection, annotation, and taxonomic classification of viral contigs in metagenomic and metatranscriptomic assemblies. The pipeline is part of the repertoire of analysis services offered by MGnify. VIRify’s taxonomic classification relies on the detection of taxon-specific profile hidden Markov models (HMMs), built upon a set of 22,014 orthologous protein domains and referred to as ViPhOGs. VIRify was implemented in CWL. What do I need? The ...
Type: Nextflow
Creators: Martin Beracochea, Martin Hölzer, Alexandre Almeida, Guillermo Rangel-Pineros and Ekaterina Sakharova
Submitter: Laura Rodriguez-Navas
VIRify
VIRify is a recently developed pipeline for the detection, annotation, and taxonomic classification of viral contigs in metagenomic and metatranscriptomic assemblies. The pipeline is part of the repertoire of analysis services offered by MGnify. VIRify’s taxonomic classification relies on the detection of taxon-specific profile hidden Markov models (HMMs), built upon a set of 22,014 orthologous protein domains and referred to as ViPhOGs. VIRify was implemented in CWL. What do I need? The ...
Type: Common Workflow Language
Creators: Martin Beracochea, Martin Hölzer, Alexandre Almeida, Guillermo Rangel-Pineros and Ekaterina Sakharova
Submitter: Laura Rodriguez-Navas
Rare disease researchers workflow is that they submit their raw data (fastq), run the mapping and variant calling RD-Connect pipeline and obtain unannotated gvcf files to further submit to the RD-Connect GPAP or analyse on their own.
This demonstrator focuses on the variant calling pipeline. The raw genomic data is processed using the RD-Connect pipeline (Laurie et al., 2016) running on the standards (GA4GH) compliant, interoperable container ...
Type: Common Workflow Language
Creators: José Mª Fernández, Laura Rodriguez-Navas
Submitter: Laura Rodriguez-Navas
nf-core/vipr is a bioinformatics best-practice analysis pipeline for assembly and intrahost / low-frequency variant calling for viral samples. The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It comes with docker / singularity containers making installation trivial and results highly reproducible. Pipeline Steps
Step Main program/s
Trimming, combining of read-pairs per sample and QC Skewer, FastQC
Decontamination ...
Type: Nextflow
Creator: Andreas Wilm and October SESSIONS and Paola Florez DE SESSIONS and ZHU Yuan and Shuzhen SIM and CHU Wenhan Collins
Submitter: Laura Rodriguez-Navas
 Download
Download