Workflow Type: Common Workflow Language
Frozen
Stable
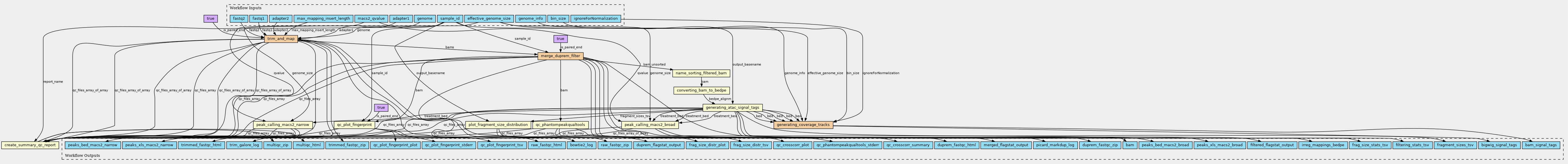
This workflow supports processing of bulk ATAC-Seq data from raw reads to genome-wide accessiblity tracks (bigWig) and ATAC peaks. The main steps include read trimming using trimGalore, alignment with bowtie2, coverage generation using samtools and peak calling with MACS2.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| sample_id | n/a | Sample ID used for naming the output files. |
|
| fastq1 | n/a | List of fastq files containing the first mate of raw reads. Muliple files are provided if multiplexing of the same library has been done on multiple lanes. The reads comming from different fastq files are pooled after alignment. Also see parameter "fastq2". |
|
| fastq2 | n/a | List of fastq files containing the second mate of raw reads. Important: this list has to be of same length as parameter "fastq1". |
|
| adapter1 | n/a | Adapter sequence for first reads. If not specified (set to "null"), trim_galore will try to autodetect whether ...\n - Illumina universal adapter (AGATCGGAAGAGC)\n - Nextera adapter (CTGTCTCTTATA)\n - Illumina Small RNA 3-prime Adapter (TGGAATTCTCGG)\n ... was used.\n You can directly choose one of the above configurations by setting the string to "illumina", "nextera", or "small_rna". Or you specify the adaptor string manually (e.g. "AGATCGGAAGAGC"). |
|
| adapter2 | n/a | Adapter sequence for second reads. If not specified (set to "null"), trim_galore will try to autodetect whether ...\n - Illumina universal adapter (AGATCGGAAGAGC)\n - Nextera adapter (CTGTCTCTTATA)\n - Illumina Small RNA 3-prime Adapter (TGGAATTCTCGG)\n ... was used.\n You can directly choose one of the above configurations by setting the string to "illumina", "nextera", or "small_rna". Or you specify the adaptor string manually (e.g. "AGATCGGAAGAGC"). |
|
| genome | n/a | Path to reference genome in fasta format. Bowtie2 index files (".1.bt2", ".2.bt2", ...) as well as a samtools index (".fai") has to be located in the same directory.\n All of these files can be downloaded for the most common genome builds at https://support.illumina.com/sequencing/sequencing_software/igenome.html. Alternatively, you can use "bowtie2-build" or "samtools index" to create them yourself. |
|
| genome_info | n/a | Path to a tab-delimited file listing chromosome sizes in following fashion:\n "chromosome_nametotal_number_of_bp".\n For the most common UCSC genome build, you can find corresponding files at: https://github.com/CompEpigen/ATACseq_workflows/tree/master/chrom_sizes. Or you can generate them yourself using UCSC script fetchChromSizes (http://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/fetchChromSizes) in following fashion:\n "fetchChromSizes hg38 > hg38.chrom.sizes".\n If you are dealing with a non-UCSC build, you can generate such a file from a samtools index using:\n "awk -v OFS='\t' {'print $1,$2'} hg38.fa.fai > hg38.chrom.sizes". |
|
| max_mapping_insert_length | n/a | Maximum insert length between two reads of a pair. In case of ATACseq, very long insert sizes are possible. So it is recommended to use at least a value of 1500. However, please note that alignment will take significantly longer for higher insert sizes. The default is 2500. |
|
| macs2_qvalue | n/a | Q-value cutoff used for peak calling by MACS2. The default is 0.05. |
|
| effective_genome_size | n/a | The effectively mappable genome size, please see: https://deeptools.readthedocs.io/en/latest/content/feature/effectiveGenomeSize.html |
|
| bin_size | n/a | Bin size used for generation of coverage tracks. The larger the bin size the smaller are the coverage tracks, however, the less precise is the signal. For single bp resolution set to 1. |
|
| ignoreForNormalization | n/a | List of space-delimited chromosome names that shall be ignored when calculating the scaling factor. Specify as space-delimited string. Default: "chrX chrY chrM" |
|
Steps
| ID | Name | Description |
|---|---|---|
| trim_and_map | n/a | n/a |
| merge_duprem_filter | n/a | n/a |
| name_sorting_filtered_bam | n/a | samtools sort - sorting of filtered bam file by read name |
| converting_bam_to_bedpe | n/a | bedtools bamtobed |
| generating_atac_signal_tags | n/a | n/a |
| generating_coverage_tracks | n/a | n/a |
| peak_calling_macs2_broad | n/a | peak calling using macs2 |
| peak_calling_macs2_narrow | n/a | peak calling using macs2 |
| plot_fragment_size_distribution | n/a | n/a |
| qc_plot_fingerprint | n/a | n/a |
| qc_phantompeakqualtools | n/a | n/a |
| create_summary_qc_report | n/a | multiqc summarizes the qc results from fastqc and other tools |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| raw_fastqc_zip | n/a | n/a |
|
| raw_fastqc_html | n/a | n/a |
|
| trim_galore_log | n/a | n/a |
|
| trimmed_fastqc_html | n/a | n/a |
|
| trimmed_fastqc_zip | n/a | n/a |
|
| bowtie2_log | n/a | n/a |
|
| duprem_fastqc_zip | n/a | n/a |
|
| duprem_fastqc_html | n/a | n/a |
|
| merged_flagstat_output | n/a | n/a |
|
| filtered_flagstat_output | n/a | n/a |
|
| duprem_flagstat_output | n/a | n/a |
|
| bam | n/a | n/a |
|
| picard_markdup_log | n/a | n/a |
|
| frag_size_stats_tsv | n/a | n/a |
|
| filtering_stats_tsv | n/a | n/a |
|
| fragment_sizes_tsv | n/a | n/a |
|
| irreg_mappings_bedpe | n/a | n/a |
|
| bam_signal_tags | n/a | n/a |
|
| bigwig_signal_tags | n/a | n/a |
|
| peaks_bed_macs2_broad | n/a | n/a |
|
| peaks_xls_macs2_broad | n/a | n/a |
|
| peaks_bed_macs2_narrow | n/a | n/a |
|
| peaks_xls_macs2_narrow | n/a | n/a |
|
| frag_size_distr_plot | n/a | n/a |
|
| frag_size_distr_tsv | n/a | n/a |
|
| qc_plot_fingerprint_plot | n/a | n/a |
|
| qc_plot_fingerprint_tsv | n/a | n/a |
|
| qc_plot_fingerprint_stderr | n/a | n/a |
|
| qc_crosscorr_summary | n/a | n/a |
|
| qc_crosscorr_plot | n/a | n/a |
|
| qc_phantompeakqualtools_stderr | n/a | n/a |
|
| multiqc_zip | n/a | n/a |
|
| multiqc_html | n/a | n/a |
|
Version History
master @ f6ad72e (earliest) Created 27th Jun 2025 at 14:53 by Pavlo Lutsik
added PDF of cwl-viewer
Frozen
 master
masterf6ad72e
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Tools
License
Activity
Views: 9 Downloads: 1
Created: 27th Jun 2025 at 14:52
Annotated Properties
 Tags
TagsThis item has not yet been tagged.
 Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub